Abstract
Ageing is accompanied by a decline in cellular proteostasis, which underlies many age-related protein misfolding diseases1,2. Yet, how ageing impairs proteostasis remains unclear. As nascent polypeptides represent a substantial burden on the proteostasis network3, we hypothesized that altered translational efficiency during ageing could help to drive the collapse of proteostasis. Here we show that ageing alters the kinetics of translation elongation in both Caenorhabditis elegans and Saccharomyces cerevisiae. Ribosome pausing was exacerbated at specific positions in aged yeast and worms, including polybasic stretches, leading to increased ribosome collisions known to trigger ribosome-associated quality control (RQC)4,5,6. Notably, aged yeast cells exhibited impaired clearance and increased aggregation of RQC substrates, indicating that ageing overwhelms this pathway. Indeed, long-lived yeast mutants reduced age-dependent ribosome pausing, and extended lifespan correlated with greater flux through the RQC pathway. Further linking altered translation to proteostasis collapse, we found that nascent polypeptides exhibiting age-dependent ribosome pausing in C. elegans were strongly enriched among age-dependent protein aggregates. Notably, ageing increased the pausing and aggregation of many components of proteostasis, which could initiate a cycle of proteostasis collapse. We propose that increased ribosome pausing, leading to RQC overload and nascent polypeptide aggregation, critically contributes to proteostasis impairment and systemic decline during ageing.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The datasets generated for this study have been deposited in NCBI’s Gene Expression Omnibus (GEO) under GEO Series accession number GSE152850. Additional datasets used in this study are also publicly available: GSE139036 (disome profiling), GSE69414 and GSE52968 (Ribo-seq of yeast treated with 3-amino-1,2,4-triazole), ref. 62 (yeast chronological lifespan), ref. 6 (RQC flux), and refs. 37 and 65 (protein aggregation during C. elegans ageing).
Code availability
All customized Python or R scripts used for data processing and analysis are available from the corresponding author on request.
References
López-Otín, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. The hallmarks of aging. Cell 153, 1194–1217 (2013).
Chiti, F. & Dobson, C. M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 75, 333–366 (2006).
Pechmann, S., Willmund, F. & Frydman, J. The ribosome as a hub for protein quality control. Mol. Cell 49, 411–421 (2013).
Simms, C. L., Yan, L. L. & Zaher, H. S. Ribosome collision is critical for quality control during no-go decay. Mol. Cell 68, 361–373 (2017).
Juszkiewicz, S. et al. ZNF598 is a quality control sensor of collided ribosomes. Mol. Cell 72, 469–481 (2018).
Brandman, O. et al. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell 151, 1042–1054 (2012).
Balchin, D., Hayer-Hartl, M. & Hartl, F. U. In vivo aspects of protein folding and quality control. Science 353, aac4354 (2016).
Jahn, T. R. & Radford, S. E. Folding versus aggregation: polypeptide conformations on competing pathways. Arch. Biochem. Biophys. 469, 100–117 (2008).
Ciryam, P., Tartaglia, G. G., Morimoto, R. I., Dobson, C. M. & Vendruscolo, M. Widespread aggregation and neurodegenerative diseases are associated with supersaturated proteins. Cell Rep. 5, 781–790 (2013).
Gingold, H. & Pilpel, Y. Determinants of translation efficiency and accuracy. Mol. Syst. Biol. 7, 481 (2011).
Stein, K. C. & Frydman, J. The stop-and-go traffic regulating protein biogenesis: how translation kinetics controls proteostasis. J. Biol. Chem. 294, 2076–2084 (2019).
Yu, C.-H. et al. Codon usage influences the local rate of translation elongation to regulate co-translational protein folding. Mol. Cell 59, 744–754 (2015).
Pechmann, S. & Frydman, J. Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding. Nat. Struct. Mol. Biol. 20, 237–243 (2013).
Kudla, G., Murray, A. W., Tollervey, D. & Plotkin, J. B. Coding-sequence determinants of gene expression in Escherichia coli. Science 324, 255–258 (2009).
Zhang, G., Hubalewska, M. & Ignatova, Z. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat. Struct. Mol. Biol. 16, 274–280 (2009).
Collart, M. A. & Weiss, B. Ribosome pausing, a dangerous necessity for co-translational events. Nucleic Acids Res. 48, 1043–1055 (2020).
Chartron, J. W., Hunt, K. C. L. & Frydman, J. Cotranslational signal-independent SRP preloading during membrane targeting. Nature 536, 224–228 (2016).
Pechmann, S., Chartron, J. W. & Frydman, J. Local slowdown of translation by nonoptimal codons promotes nascent-chain recognition by SRP in vivo. Nat. Struct. Mol. Biol. 21, 1100–1105 (2014).
Stein, K. C., Kriel, A. & Frydman, J. Nascent polypeptide domain topology and elongation rate direct the cotranslational hierarchy of Hsp70 and TRiC/CCT. Mol. Cell 75, 1117–1130 (2019).
Sitron, C. S. & Brandman, O. Detection and degradation of stalled nascent chains via ribosome-associated quality control. Annu. Rev. Biochem. 89, 417–442 (2020).
Brandman, O. & Hegde, R. S. Ribosome-associated protein quality control. Nat. Struct. Mol. Biol. 23, 7–15 (2016).
Dimitrova, L. N., Kuroha, K., Tatematsu, T. & Inada, T. Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. J. Biol. Chem. 284, 10343–10352 (2009).
Buhr, F. et al. Synonymous codons direct cotranslational folding toward different protein conformations. Mol. Cell 61, 341–351 (2016).
Nedialkova, D. D. & Leidel, S. A. Optimization of codon translation rates via trna modifications maintains proteome integrity. Cell 161, 1606–1618 (2015).
Kim, S. J. et al. Protein folding. Translational tuning optimizes nascent protein folding in cells. Science 348, 444–448 (2015).
Willmund, F. et al. The cotranslational function of ribosome-associated Hsp70 in eukaryotic protein homeostasis. Cell 152, 196–209 (2013).
Duttler, S., Pechmann, S. & Frydman, J. Principles of cotranslational ubiquitination and quality control at the ribosome. Mol. Cell 50, 379–393 (2013).
Koplin, A. et al. A dual function for chaperones SSB-RAC and the NAC nascent polypeptide-associated complex on ribosomes. J. Cell Biol. 189, 57–68 (2010).
Choe, Y.-J. et al. Failure of RQC machinery causes protein aggregation and proteotoxic stress. Nature 531, 191–195 (2016).
Yonashiro, R. et al. The Rqc2/Tae2 subunit of the ribosome-associated quality control (RQC) complex marks ribosome-stalled nascent polypeptide chains for aggregation. eLife 5, e11794 (2016).
Wu, C. C.-C., Peterson, A., Zinshteyn, B., Regot, S. & Green, R. Ribosome collisions trigger general stress responses to regulate cell fate. Cell 182, 404–416 (2020).
Ishimura, R. et al. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science 345, 455–459 (2014).
Bengtson, M. H. & Joazeiro, C. A. P. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature 467, 470–473 (2010).
Taylor, R. C. & Dillin, A. Aging as an event of proteostasis collapse. Cold Spring Harb. Perspect. Biol. 3, a004440 (2011).
Ben-Zvi, A., Miller, E. A. & Morimoto, R. I. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc. Natl Acad. Sci. USA 106, 14914–14919 (2009).
Steffen, K. K. & Dillin, A. A ribosomal perspective on proteostasis and aging. Cell Metab. 23, 1004–1012 (2016).
Walther, D. M. et al. Widespread proteome remodeling and aggregation in aging C. elegans. Cell 161, 919–932 (2015).
Pan, K. Z. et al. Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell 6, 111–119 (2007).
Hansen, M. et al. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell 6, 95–110 (2007).
Reis-Rodrigues, P. et al. Proteomic analysis of age-dependent changes in protein solubility identifies genes that modulate lifespan. Aging Cell 11, 120–127 (2012).
Narayan, V. et al. Deep proteome analysis identifies age-related processes in C. elegans. Cell Syst. 3, 144–159 (2016).
Hu, Z. et al. Ssd1 and Gcn2 suppress global translation efficiency in replicatively aged yeast while their activation extends lifespan. eLife 7, 4443 (2018).
Young, D. J., Guydosh, N. R., Zhang, F., Hinnebusch, A. G. & Green, R. Rli1/ABCE1 recycles terminating ribosomes and controls translation reinitiation in 3'UTRs in vivo. Cell 162, 872–884 (2015).
Guydosh, N. R. & Green, R. Dom34 rescues ribosomes in 3' untranslated regions. Cell 156, 950–962 (2014).
Choi, J. et al. How messenger RNA and nascent chain sequences regulate translation elongation. Annu. Rev. Biochem. 87, 421–449 (2018).
Schuller, A. P., Wu, C. C.-C., Dever, T. E., Buskirk, A. R. & Green, R. eIF5A functions globally in translation elongation and termination. Mol. Cell 66, 194–205 (2017).
Meydan, S. & Guydosh, N. R. Disome and trisome profiling reveal genome-wide targets of ribosome quality control. Mol. Cell 79, 588–602 (2020).
Gamble, C. E., Brule, C. E., Dean, K. M., Fields, S. & Grayhack, E. J. Adjacent codons act in concert to modulate translation efficiency in yeast. Cell 166, 679–690 (2016).
Han, P. et al. Genome-wide survey of ribosome collision. Cell Rep. 31, 107610 (2020).
Juszkiewicz, S., Speldewinde, S. H., Wan, L., Svejstrup, J. Q. & Hegde, R. S. The ASC-1 complex disassembles collided ribosomes. Mol. Cell 79, 603–614 (2020).
Ikeuchi, K. et al. Collided ribosomes form a unique structural interface to induce Hel2-driven quality control pathways. EMBO J. 38, e100276 (2019).
Shen, P. S. et al. Protein synthesis. Rqc2p and 60S ribosomal subunits mediate mRNA-independent elongation of nascent chains. Science 347, 75–78 (2015).
Shao, S., Malsburg, von der, K. & Hegde, R. S. Listerin-dependent nascent protein ubiquitination relies on ribosome subunit dissociation. Mol. Cell 50, 637–648 (2013).
Shao, S. & Hegde, R. S. Reconstitution of a minimal ribosome-associated ubiquitination pathway with purified factors. Mol. Cell 55, 880–890 (2014).
Shao, S., Brown, A., Santhanam, B. & Hegde, R. S. Structure and assembly pathway of the ribosome quality control complex. Mol. Cell 57, 433–444 (2015).
Juszkiewicz, S. & Hegde, R. S. Initiation of quality control during poly(A) translation requires site-specific ribosome ubiquitination. Mol. Cell 65, 743–750 (2017).
Sundaramoorthy, E. et al. ZNF598 and RACK1 regulate mammalian ribosome-associated quality control function by mediating regulatory 40S ribosomal ubiquitylation. Mol. Cell 65, 751–760 (2017).
Matsuo, Y. et al. Ubiquitination of stalled ribosome triggers ribosome-associated quality control. Nat. Commun. 8, 159 (2017).
Tsuboi, T. et al. Dom34:hbs1 plays a general role in quality-control systems by dissociation of a stalled ribosome at the 3′ end of aberrant mRNA. Mol. Cell 46, 518–529 (2012).
Sitron, C. S. & Brandman, O. CAT tails drive on- and off-ribosome degradation of stalled polypeptides. Nat. Struct. Mol. Biol. 26, 450–459 (2018).
Anisimova, A. S. et al. Multifaceted deregulation of gene expression and protein synthesis with age. Proc. Natl Acad. Sci. USA 117, 15581–15590 (2020).
Powers, R. W., Kaeberlein, M., Caldwell, S. D., Kennedy, B. K. & Fields, S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 20, 174–184 (2006).
Fabrizio, P., Pozza, F., Pletcher, S. D., Gendron, C. M. & Longo, V. D. Regulation of longevity and stress resistance by Sch9 in yeast. Science 292, 288–290 (2001).
Kaeberlein, M. et al. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science 310, 1193–1196 (2005).
David, D. C. et al. Widespread protein aggregation as an inherent part of aging in C. elegans. PLoS Biol. 8, e1000450 (2010).
Barros, G. C. et al. Rqc1 and other yeast proteins containing highly positively charged sequences are not targets of the RQC complex. J. Biol. Chem. 296, 100586 (2021).
Brenner, S. The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974).
Winzeler, E. A. et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906 (1999).
Yofe, I. et al. One library to make them all: streamlining the creation of yeast libraries via a SWAp-Tag strategy. Nat. Chem. Biol. 13, 371–378 (2016).
Huh, W.-K. et al. Global analysis of protein localization in budding yeast. Nature 425, 686–691 (2003).
Hughes, A. L. & Gottschling, D. E. An early age increase in vacuolar pH limits mitochondrial function and lifespan in yeast. Nature 492, 261–265 (2012).
Ingolia, N. T., Brar, G. A., Rouskin, S., McGeachy, A. M. & Weissman, J. S. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 7, 1534–1550 (2012).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Dey, K. K., Xie, D. & Stephens, M. A new sequence logo plot to highlight enrichment and depletion. BMC Bioinform. 19, 473 (2018).
Burtner, C. R., Murakami, C. J., Kennedy, B. K. & Kaeberlein, M. A molecular mechanism of chronological aging in yeast. Cell Cycle 8, 1256–1270 (2009).
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
McCormick, M. A. et al. A comprehensive analysis of replicative lifespan in 4,698 single-gene deletion strains uncovers conserved mechanisms of aging. Cell Metab. 22, 895–906 (2015).
Acknowledgements
We thank R. Aviner and M. Aguilar-Rangel for comments on the manuscript; members of the Frydman Laboratory for discussions and advice; J. Chartron, P. Dolan and K. Dalton for technical input; and J. Lim and J. Arribere for C. elegans expertise. YTM1 and HAT2performed at the UCSF Center for Advanced Technology. K.C.S. was supported as a Glenn Foundation for Medical Research Postdoctoral Fellow and by NIH/NIA grant (T32AG047126). F.M.-P. was supported by The Pew Trusts in the Biomedical Sciences postdoctoral Award (00034104), and T.K.R was supported by the NIH/NIGMS National Research Service Award (F32GM120947). This work was supported by NIH grants GM056433 and AG054407 (to J.F.). J.F. is a CZ Biohub Investigator.
Author information
Authors and Affiliations
Contributions
K.C.S. and J.F. designed the study. K.C.S. performed all of the experiments and computational analyses, with assistance from F.M.-P., J.L. and T.K.R. in carrying out the yeast reporter immunoblot and microscopy experiments. K.C.S. and J.F. wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review information
Nature thanks Andrew Dillin, Toshifumi Inada and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
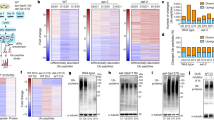
Extended Data Fig. 1 Decreased protein synthesis and translatome changes during chronological ageing of yeast.
a, Survival curve of yeast during chronological ageing. n = 3 biological replicates with mean ± s.e.m. b – c, Average ribosome occupancy at the b, start and c, stop codons during yeast chronological ageing. The shaded region represents the 95% bootstrapped confidence interval. d, Polysome profiles of chronologically aged yeast at the indicated ages showing decreased population of polysomes during ageing. n ≥ 3 biological replicates with representative profiles shown. e, Heat map of gene expression analysis of Ribo-seq data showing Pearson’s correlation coefficient between all yeast samples. f, Volcano plot of differential gene expression shows widespread translatome changes for chronologically aged yeast. n = 2,981 total genes with 901 genes having increased ribosome occupancy (purple, > 2 fold up, adjusted P < 0.05, Benjamini-Hochberg method) and 1,033 genes having decreased ribosome occupancy (green, > 2 fold down, adjusted P < 0.05, Benjamini-Hochberg method) in Day 4 yeast cells relative to Day 0 yeast cells. g, Functional enrichment of differentially translated genes during ageing displaying representative gene ontology terms (adjusted P < 0.1, Benjamini-Hochberg method). See Supplementary Table 2 for complete, unfiltered results. h, Ribosome occupancy on ribosomal protein RPL3 showing decreased translation during yeast ageing. i, Ribosome occupancy on the 5’UTR and coding sequence of GCN4, showing decreased occupancy at upstream ORFs and increased occupancy across the coding sequence during ageing. j, Scatter plot of average pause scores for each amino acid residue in coding sequences of young Day 0 and aged Day 4 yeast. k, Scatter plots of average pause scores for each amino acid residue in coding sequences of two biological replicates of Day 0 (left) and Day 4 (right) chronologically aged yeast.
Extended Data Fig. 2 Decreased protein synthesis and translatome changes during chronological ageing of worms.
a, Polysome profiles of adult worms at the indicated ages showing decreased population of polysomes during ageing. n ≥ 3 biological replicates with representative profiles shown. b – c, Average ribosome occupancy at the b, start and c, stop codons of young Day 1 and aged Day 12 worms. The shaded region represents the 95% bootstrapped confidence interval. d, Heat map of gene expression analysis of Ribo-seq data showing Pearson’s correlation coefficient between worm samples. e, Volcano plot of differential gene expression shows widespread translatome changes during worm ageing. n = 8,341 total genes with 621 genes having increased ribosome occupancy (purple, > 2 fold up, adjusted P < 0.05, Benjamini-Hochberg method) and 655 genes having decreased ribosome occupancy (green, > 2 fold down, adjusted P < 0.05, Benjamini-Hochberg method) in Day 12 adult worms relative to Day 1 adult worms. f, Functional enrichment of differentially translated genes displaying representative gene ontology terms (adjusted P < 0.1, except for categories with number of genes < 20 where adjusted P < 0.25, Benjamini-Hochberg method). See Supplementary Table 2 for complete, unfiltered results. g, Ribosome occupancy on ribosomal protein rps-3 showing decreased translation during ageing of worms. h, Scatter plot of average pause scores for each amino acid residue in coding sequences of Day 1 and Day 12 adult worms. i, Scatter plots of average pause scores for each amino acid residue in coding sequences of two biological replicates of Day 1 (left) and Day 12 (right) adult worms.
Extended Data Fig. 3 Age-dependent ribosome pausing.
a, Scatter plot of the average pause score for each amino acid residue in coding sequences of young WT yeast cells either untreated or treated with 3-amino-1,2,4-triazole (3-AT) from previously published Ribo-seq data43. b, Amino acid residue frequency in the ribosomal A-site of statistically significant ribosome pause sites enriched in yeast cells treated with 3-AT43, relative to the residue frequency in the transcriptome. c, Functional enrichment of genes with an age-dependent pause site in yeast displaying representative gene ontology terms (adjusted P < 0.1, Benjamini–Hochberg method). See Supplementary Table 2 for complete, unfiltered results. d, Functional enrichment of genes with an age-dependent pause site in worms displaying representative gene ontology terms (adjusted P < 0.1, Benjamini–Hochberg method). See Supplementary Table 2 for complete, unfiltered results. e, Investigating the association between age-dependent ribosome pausing and co-translational ubiquitination27. Population n = 937 transcripts with a pause site, 3,145 transcripts without a pause site. P = 0.002, two-sided Fisher’s exact test. f, Codon frequency in the ribosomal A-site of age-dependent ribosome pause sites relative to the codon frequency in the transcriptome. n = 271 pause sites with Day 4 pause score > 6 in 232 genes. g – i, Peptide motif associated with greater ribosome pausing in the indicated age comparisons.
Extended Data Fig. 4 Association of age-dependent ribosome pausing in yeast with disome formation.
a, Peptide motif associated with disome formation in young cells47. b, Ribosome occupancy on HAT2 with age-dependent ribosome pausing at position 188 with Trp in the A-site. c, Occupancy of monosomes and disomes on HAT2 from disome footprint profiling of young cells47. d – h, Average ribosome occupancy in young and aged yeast, with a lagging ribosome peak present in aged cells, at d, the position where disomes were most enriched in each transcript in young cells47; e, inhibitory codon pairs previously associated with ribosome pausing48 and disome formation47; and f – h, tripeptide motifs previously associated with disome formation47. The shaded region represents the 95% bootstrapped confidence interval.
Extended Data Fig. 5 Age-dependent ribosome pausing at polybasic regions.
a – b, Average ribosome occupancy at polybasic regions of a, monosomes and b, disomes in young WT yeast47 showing length-dependent pausing and collisions. c, Average ribosome occupancy at polybasic regions consisting of 3 (left) or 5 (right) consecutive Lys or Arg (K/R) for young Day 0 and aged Day 4 yeast. n = 3,729 sites of 3 K/R in 2,198 genes, and 159 sites of 5 K/R in 152 genes. The shaded region represents the 95% bootstrapped confidence interval. Arrow indicates a putative increase in ribosome collision events with age at regions with 5 K/R. d – e, Average ribosome occupancy at polybasic regions of d, young Day 0 and e, aged Day 4 yeast. Also see panel c and Fig. 2c and 2d. f, Ribosome occupancy on SIS1 with inset highlighting ribosome pausing and collision (arrow) at stretch of 6 K/R within a 10 residue window.
Extended Data Fig. 6 Age-dependent aberrant translation and aggregation of truncated nascent polypeptides in yeast.
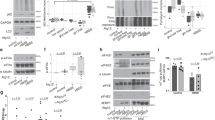
a, Immunoblot of young Day 0 and aged Day 4 yeast of the indicated strains harboring the no-insert GFP-RFP control reporter. Truncated and full-length products are noted. n ≥ 3 biological replicates with representative example shown. For gel source data, see Supplementary Fig. 1. b, Increased formation of truncated polypeptides in chronologically aged yeast is not due to acidification of the media or respiratory conditions. Immunoblot of young Day 0 and aged Day 4 WT yeast harboring the GFP-R12-FLAG-HIS3 ribosome pausing reporter and grown in standard media containing 2% glucose as carbon source, media buffered to pH 6.0, or media containing 3% glycerol. n ≥ 3 biological replicates with representative example shown. For gel source data, see Supplementary Figure 2. c, Depletion of media nutrients during yeast chronological ageing does not increase formation of truncated polypeptides. Immunoblot of GFP-K12-FLAG-HIS3 ribosome pausing reporter from young Day 0 and aged Day 4 WT or ltn1Δ yeast, as well as after swapping media (i.e. young Day 0 yeast cells grown in “aged” media from Day 4 cells). n ≥ 3 biological replicates with representative example shown. For gel source data, see Supplementary Fig. 2. d – f, Fluorescence microscopy examining puncta formation in young (Y) Day 0 and aged (A) Day 4 yeast of WT and RQC mutant cells harboring d, GFP-RFP, or e, the ribosome pausing reporter with 12 K inserted between GFP and RFP. n ≥ 3 biological replicates with representative examples shown. Scale bar: 3 µm. f, Associated quantification of GFP+RFP− puncta from n ≥ 200 cells from 3 biological replicates with mean ± SEM. For cells harboring the K12 reporter: WT: P = 2.2e−4, ltn1Δ: P = 1.3e−4; rqc2Δ: P = 9.6e−5, hel2Δ: P = 1.7e−6, two-sided Welch’s t-test. g, Immunoblot of young Day 0 and aged Day 4 ltn1Δ yeast harboring the GFP-K12-FLAG-HIS3 ribosome pausing reporter and probed with αGFP and αFLAG antibodies. Truncated, full-length, and high-molecular weight (HMW) species are noted. n ≥ 3 biological replicates with representative example shown. An asterisk around 50 kDa indicates a non-specific band from αFLAG antibody. For gel source data, see Supplementary Fig. 2.
Extended Data Fig. 7 Age-dependent ribosome pausing, aberrant translation, and aggregation of endogenous yeast proteins and reporters.
a, Immunoblot of young Day 0 and aged Day 4 yeast harboring GFP-tagged YTM1 that is either N-terminally tagged to monitor co-translational products or C-terminally tagged to monitor post-translational degradation products. n = 3 biological replicates with representative example shown. For gel source data, see Supplementary Fig. 3. b, Fluorescence microscopy and quantification of GFP+ puncta of GFP-tagged YTM1 in young and aged WT and ltn1Δ yeast cells. n ≥ 200 cells from 3 biological replicates with representative examples shown and mean ± s.e.m. WT N-term: P = 2.0e−4; ltn1Δ N-term: P = 2.3e−5; ltn1Δ C-term: P = 2.8e−4, two-sided Welch’s t-test. Scale bar: 3 µm. c, Immunoblot of young Day 0 and aged Day 4 yeast harboring N-terminally GFP-tagged HAT2 showing increased truncated co-translational products during ageing. n = 3 biological replicates with representative example shown. For gel source data, see Supplementary Fig. 3. d, Quantification of GFP+RFP+ puncta in hel2Δ cells using the indicated reporter. n ≥ 200 cells from 3 biological replicates with mean ± s.e.m. K12: P = 7.0e−4; R12: P = 2.5e−4, two-sided Welch’s t-test. e, Ribosome occupancy across the stalling reporter GFP-K12-FLAG-HIS3 (left) and the no-insert control (right) of young Day 0 and aged Day 4 yeast, showing increased occupancy in 3’ end of the stalling reporter coding sequence during ageing.
Extended Data Fig. 8 Association of extended lifespan with improved RQC flux and reduced ribosome pausing and collision.
a, RQC flux was assessed based on the abundance of the GFP-R12-HIS3 ribosome pausing reporter6. RQC flux of the following four groups of genes was compared to yeast strains with deficient RQC (i.e. strains with reporter GFP abundance greater than 3 standard deviations above the mean of the total collection;6 n = 39): 1) long-lived CLS strains (strains with a chronological lifespan greater than 1 standard deviation above the mean of the total collection;62 n = 150, P = 7.3e−22), 2) genes involved in the TOR pathway (n = 31, P = 2.8e−20), 3) long-lived CLS strains not involved in the TOR pathway (n = 142, P = 9.3e−23), and 4) long-lived RLS strains78 (n = 201, P = 1.3e−21). Statistical analysis was performed using two-sided Wilcoxon rank-sum tests. b, Heat map of gene expression analysis of Ribo-seq data showing Pearson’s correlation coefficient between all yeast samples. c, Average ribosome occupancy at age-dependent pause sites (n = 5,951 sites in 937 genes) in the indicated strains, with the distribution of pause scores in aged (A) Day 4 WT and sch9∆ yeast cells on the right (P = 2.5e−77, two-sided Wilcoxon rank-sum test). d, Peptide motif associated with positions in the translatome where ribosome pausing is greater in aged Day 4 WT yeast relative to aged Day 4 sch9∆ yeast. e, Average ribosome occupancy in WT and sch9∆ yeast at polybasic regions consisting of 6 consecutive Lys or Arg (K/R). n = 60 K/R sites in 58 genes. The shaded region represents the 95% bootstrapped confidence interval. Arrow indicates a putative increase in ribosome collision events. f, Ribosome occupancy on HAT2 (top) and YTM1 (bottom) in the indicated yeast strains around age-dependent ribosome pause sites (HAT2: position 188 with Trp in the A-site; YTM1: a stretch of 5 consecutive K/R). g, Fluorescence microscopy of GFP+RFP− puncta formation in young (Y) Day 0 and aged (A) Day 4 yeast of WT and sch9∆ cells harboring the indicated reporter. n ≥ 3 biological replicates with representative examples shown. Scale bar: 3 um. h, Immunoblot of young Day 0 and aged Day 4 yeast of WT or sch9Δ cells harboring the indicated reporter. Truncated and full-length products are noted. n ≥ 3 biological replicates with representative example shown. For gel source data, see Supplementary Fig. 4. i, Relative gene expression of RQC machinery in the translatome of WT or sch9Δ cells comparing aged Day 4 yeast to young Day 0 yeast. ns = not significant; WT / sch9Δ cells: ASC1: P = 4.1e−40 / 9.8e−26, HEL2: P = 0.002 / 1.3e−4, LTN1: P = 0.006 / ns, RQC2: P = 1.2e−9 / 2.0e−20, two-sided Wald test with Benjamini-Hochberg correction.
Extended Data Fig. 9 Sequence specificity of age-dependent ribosome pausing in worms.
a, Amino acid residue frequency in the ribosomal P- and E-sites of age-dependent ribosome pause sites relative to the residue frequency in the transcriptome. n = 587 pause sites with Day 12 pause score > 10 in 437 genes. P-site Arg: P = 0.01, E-site Arg: P = 0.04, two-sided Fisher’s exact test. b, Codon frequency in the ribosomal A-site of age-dependent ribosome pause sites relative to the codon frequency in the transcriptome. n = 587 pause sites with Day 12 pause score > 10 in 437 genes. c – e, Peptide motif associated with greater ribosome pausing in the indicated age comparisons. f, Ribosome occupancy on K12C11.6 with ribosome pausing at position 62 with Trp in the ribosomal active site. g, Average ribosome occupancy at polybasic regions consisting of 4 (left) or 5 (right) consecutive Lys or Arg (K/R). n = 1,396 sites of 4 K/R in 1,720 genes, and 369 sites of 5 K/R in 348 genes. The shaded region represents the 95% bootstrapped confidence interval.
Extended Data Fig. 10 Association of age-dependent aggregation and ribosome pausing in worms.
a, Investigating the association between age-dependent ribosome pausing and aggregation65. Population n = 742 proteins with pause site, 6,219 proteins without a pause site. P = 1.6e−23, two-sided Fisher’s exact test. b, Enrichment of polybasic regions (4 or more consecutive Lys or Arg, and 2 or more consecutive Arg) in age-dependent aggregated proteins from two different datasets37,65, and those proteins that also have age-dependent ribosome pausing. Walther, et al.: 4+ Lys/Arg: P = 0.01 (aggregate) and P = 0.03 (pause + agg.), 2+ Arg: P = 4.9e−6 (aggregate) and P = 4.8e−5 (pause + agg.); David, et al.: 4+ Lys/Arg: P = 0.05 (aggregate) and P = 0.04 (pause + agg.), 2+ Arg: P = 0.02 (aggregate) and P = 0.01 (pause + agg.), two-sided Fisher’s exact test. c, Comparing the functional enrichment within aggregated proteins65 to those that also have age-dependent ribosome pausing, displaying representative gene ontology terms (adjusted P < 0.05, Benjamini-Hochberg correction). Categories related to proteostasis are highlighted. Also see Supplementary Table 2. d, Aggregate abundance37 over time of tRNA synthetases exhibiting both age-dependent aggregation and ribosome pausing. e, Relative gene expression of worm RQC orthologs (yeast ortholog in parentheses) in the translatome in aged Day 12 adult worms compared to young Day 1 worms. ns = not significant, K04D7.1: P = 1.2e−14, K07A12.1: P = 3.9e−18, Y82E9BR.18: P = 5.4e−7, two-sided Wald test with Benjamini-Hochberg correction. f, Aggregate abundance37 over time of worm RQC orthologs.
Supplementary information
Supplementary Information
Supplementary Discussion, elaborating on the implications of our study, and Supplementary Figs. 1–4, showing the uncropped source data of our immunoblotting experiments.
Supplementary Table 1
Age-dependent pause sites in worms and yeast. The age-dependent pause sites that were identified as having a statistically significant increase in ribosome pausing during ageing.
Supplementary Table 2
Gene Ontology analysis. The unfiltered gene ontology terms associated with (1) differential gene expression during ageing; (2) genes containing an age-dependent pause site; and (3) worm genes associated with age-dependent protein aggregation.
Supplementary Table 3
Polybasic regions in worms and yeast. The regions of 3, 4, 5 or 6+ consecutive Lys (K) or Arg (R) residues identified in coding sequences.
Rights and permissions
About this article
Cite this article
Stein, K.C., Morales-Polanco, F., van der Lienden, J. et al. Ageing exacerbates ribosome pausing to disrupt cotranslational proteostasis. Nature 601, 637–642 (2022). https://doi.org/10.1038/s41586-021-04295-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-04295-4
This article is cited by
-
Transcriptional profile of ribosome-associated quality control components and their associated phenotypes in mammalian cells
Scientific Reports (2024)
-
Riboformer: a deep learning framework for predicting context-dependent translation dynamics
Nature Communications (2024)
-
Transcriptional characteristics and functional validation of three monocyte subsets during aging
Immunity & Ageing (2023)
-
Mechanisms and pathology of protein misfolding and aggregation
Nature Reviews Molecular Cell Biology (2023)
-
Ribosome biogenesis in disease: new players and therapeutic targets
Signal Transduction and Targeted Therapy (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.