Abstract
The abnormal aggregation of TAR DNA-binding protein 43 kDa (TDP-43) in neurons and glia is the defining pathological hallmark of the neurodegenerative disease amyotrophic lateral sclerosis (ALS) and multiple forms of frontotemporal lobar degeneration (FTLD)1,2. It is also common in other diseases, including Alzheimer’s and Parkinson’s. No disease-modifying therapies exist for these conditions and early diagnosis is not possible. The structures of pathological TDP-43 aggregates are unknown. Here we used cryo-electron microscopy to determine the structures of aggregated TDP-43 in the frontal and motor cortices of an individual who had ALS with FTLD and from the frontal cortex of a second individual with the same diagnosis. An identical amyloid-like filament structure comprising a single protofilament was found in both brain regions and individuals. The ordered filament core spans residues 282–360 in the TDP-43 low-complexity domain and adopts a previously undescribed double-spiral-shaped fold, which shows no similarity to those of TDP-43 filaments formed in vitro3,4. An abundance of glycine and neutral polar residues facilitates numerous turns and restricts β-strand length, which results in an absence of β-sheet stacking that is associated with cross-β amyloid structure. An uneven distribution of residues gives rise to structurally and chemically distinct surfaces that face external densities and suggest possible ligand-binding sites. This work enhances our understanding of the molecular pathogenesis of ALS and FTLD and informs the development of diagnostic and therapeutic agents that target aggregated TDP-43.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Cryo-EM datasets have been deposited to the Electron Microscopy Public Image Archive (EMPIAR) under the following accession numbers: 10830 for individual one, frontal cortex; 10831 for individual one, motor cortex; and 10832 for individual two, frontal cortex. Cryo-EM maps have been deposited to the Electron Microscopy Data Bank (EMDB) under the following accession numbers: 13708 for individual one, frontal cortex; 13710 for individual one, motor cortex; and 13712 for individual two, frontal cortex. The atomic model of the double-spiral fold has been deposited to the Protein Data Bank (PDB) under accession number 7PY2. The atomic model of the α-subunit of glutamate synthase was obtained from the PDB (accession number 1EA0). The atomic model of the α-helices formed in solution by the hydrophobic region of TDP-43 was obtained from the PDB (accession number 2N3X). The atomic models of TDP-43 filaments formed in vitro were obtained from the PDB (accession numbers 7KWZ, 6N3C, 6N3A, 6N37 and 6N3B). Mass spectrometry data have been deposited to the Japan Proteome Standard Repository (jPOSTrepo) under accession number PXD029001. Any other data are available from the corresponding author upon request.
References
Neumann, M. et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133 (2006).
Arai, T. et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 351, 602–611 (2006).
Cao, Q., Boyer, D. R., Sawaya, M. R., Ge, P. & Eisenberg, D. S. Cryo-EM structures of four polymorphic TDP-43 amyloid cores. Nat. Struct. Mol. Biol. 26, 619–627 (2019).
Li, Q., Babinchak, W. M. & Surewicz, W. K. Cryo-EM structure of amyloid fibrils formed by the entire low complexity domain of TDP-43. Nat. Commun. 12, 1620 (2021).
Brettschneider, J. et al. Stages of pTDP-43 pathology in amyotrophic lateral sclerosis. Ann. Neurol. 74, 20–38 (2013).
Nonaka, T. et al. Prion-like properties of pathological TDP-43 aggregates from diseased brains. Cell Rep. 4, 124–134 (2013).
Phukan, J., Pender, N. P. & Hardiman, O. Cognitive impairment in amyotrophic lateral sclerosis. Lancet Neurol. 6, 994–1003 (2007).
Sreedharan, J. et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 319, 1668–1672 (2008).
Kabashi, E. et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat. Genet. 40, 572–574 (2008).
Van Deerlin, V. M. et al. TARDBP mutations in amyotrophic lateral sclerosis with TDP-43 neuropathology: a genetic and histopathological analysis. Lancet Neurol. 7, 409–416 (2008).
Hergesheimer, R. C. et al. The debated toxic role of aggregated TDP-43 in amyotrophic lateral sclerosis: a resolution in sight? Brain 142, 1176–1194 (2019).
Ayala, Y. M. et al. Structural determinants of the cellular localization and shuttling of TDP-43. J. Cell Sci. 121, 3778–3785 (2008).
Afroz, T. et al. Functional and dynamic polymerization of the ALS-linked protein TDP-43 antagonizes its pathologic aggregation. Nat. Commun. 8, 45 (2017).
Colombrita, C. et al. TDP-43 is recruited to stress granules in conditions of oxidative insult. J. Neurochem. 111, 1051–1061 (2009).
Hasegawa, M. et al. Phosphorylated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Ann. Neurol. 64, 60–70 (2008).
Lin, W.-L. & Dickson, D. W. Ultrastructural localization of TDP-43 in filamentous neuronal inclusions in various neurodegenerative diseases. Acta Neuropathol. 116, 205–213 (2008).
Mori, F. et al. Maturation process of TDP-43-positive neuronal cytoplasmic inclusions in amyotrophic lateral sclerosis with and without dementia. Acta Neuropathol. 116, 193–203 (2008).
Tsuji, H. et al. Molecular analysis and biochemical classification of TDP-43 proteinopathy. Brain 135, 3380–3391 (2012).
Mackenzie, I. R. A. et al. A harmonized classification system for FTLD-TDP pathology. Acta Neuropathol. 122, 111–113 (2011).
Jiang, L.-L. et al. Structural transformation of the amyloidogenic core region of TDP-43 protein initiates its aggregation and cytoplasmic inclusion. J. Biol. Chem. 288, 19614–19624 (2013).
Kametani, F. et al. Mass spectrometric analysis of accumulated TDP-43 in amyotrophic lateral sclerosis brains. Sci. Rep. 6, 23281 (2016).
Sunde, M. et al. Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J. Mol. Biol. 273, 729–739 (1997).
Binda, C. et al. Cross-talk and ammonia channeling between active centers in the unexpected domain arrangement of glutamate synthase. Structure 8, 1299–1308 (2000).
Parkinson, G. N., Cuenca, F. & Neidle, S. Topology conservation and loop flexibility in quadruplex–drug recognition: crystal structures of inter- and intramolecular telomeric DNA quadruplex–drug complexes. J. Mol. Biol. 381, 1145–1156 (2008).
Shi, Y. et al. Structure-based classification of tauopathies. Nature 598, 359–363 (2021).
Schweighauser, M. et al. Structures of α-synuclein filaments from multiple system atrophy. Nature 585, 464–469 (2020).
Kollmer, M. et al. Cryo-EM structure and polymorphism of Aβ amyloid fibrils purified from Alzheimer’s brain tissue. Nat. Commun. 10, 4760 (2019).
Krebs, M. R. H. et al. The binding of thioflavin-T to amyloid fibrils: localisation and implications. J. Struct. Biol. 149, 30–37 (2005).
Shi, Y. et al. Cryo-EM structures of tau filaments from Alzheimer’s disease with PET ligand APN-1607. Acta Neuropathol. 141, 697–708 (2021).
Bigio, E. H. et al. Inclusions in frontotemporal lobar degeneration with TDP-43 proteinopathy (FTLD-TDP) and amyotrophic lateral sclerosis (ALS), but not FTLD with FUS proteinopathy (FTLD-FUS), have properties of amyloid. Acta Neuropathol. 125, 463–465 (2013).
Robinson, J. L. et al. TDP-43 skeins show properties of amyloid in a subset of ALS cases. Acta Neuropathol. 125, 121–131 (2013).
Sun, Y. et al. Physiologically important electrolytes as regulators of TDP-43 aggregation and droplet-phase behavior. Biochemistry 58, 590–607 (2019).
Marquié, M. et al. Validating novel tau positron emission tomography tracer [F-18]-AV-1451 (T807) on postmortem brain tissue. Ann. Neurol. 78, 787–800 (2015).
Lowe, V. J. et al. An autoradiographic evaluation of AV-1451 tau PET in dementia. Acta Neuropathol. Commun. 4, 58 (2016).
Sander, K. et al. Characterization of tau positron emission tomography tracer [18F]AV-1451 binding to postmortem tissue in Alzheimer’s disease, primary tauopathies, and other dementias. Alzheimers Dement. 12, 1116–1124 (2016).
Zhang, W. et al. Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer’s and Pick’s diseases. eLife 8, e43584 (2019).
Murray, D. T. et al. Structure of FUS protein fibrils and its relevance to self-assembly and phase separation of low-complexity domains. Cell 171, 615-627.e16 (2017).
Lu, J. et al. CryoEM structure of the low-complexity domain of hnRNPA2 and its conversion to pathogenic amyloid. Nat. Commun. 11, 4090 (2020).
Sun, Y. et al. The nuclear localization sequence mediates hnRNPA1 amyloid fibril formation revealed by cryoEM structure. Nat. Commun. 11, 6349 (2020).
Lee, M., Ghosh, U., Thurber, K. R., Kato, M. & Tycko, R. Molecular structure and interactions within amyloid-like fibrils formed by a low-complexity protein sequence from FUS. Nat. Commun. 11, 5735 (2020).
Chen, A. K.-H. et al. Induction of amyloid fibrils by the C-terminal fragments of TDP-43 in amyotrophic lateral sclerosis. J. Am. Chem. Soc. 132, 1186–1187 (2010).
Guo, W. et al. An ALS-associated mutation affecting TDP-43 enhances protein aggregation, fibril formation and neurotoxicity. Nat. Struct. Mol. Biol. 18, 822–830 (2011).
Laferrière, F. et al. TDP-43 extracted from frontotemporal lobar degeneration subject brains displays distinct aggregate assemblies and neurotoxic effects reflecting disease progression rates. Nat. Neurosci. 22, 65–77 (2019).
Porta, S. et al. Patient-derived frontotemporal lobar degeneration brain extracts induce formation and spreading of TDP-43 pathology in vivo. Nat. Commun. 9, 4220 (2018).
Yoshida, M. Amyotrophic lateral sclerosis with dementia: the clinicopathological spectrum. Neuropathology 24, 87–102 (2004).
Taniguchi-Watanabe, S. et al. Biochemical classification of tauopathies by immunoblot, protein sequence and mass spectrometric analyses of Sarkosyl-insoluble and trypsin-resistant tau. Acta Neuropathol. 131, 267–280 (2016).
Tsuji, H. et al. Epitope mapping of antibodies against TDP-43 and detection of protease-resistant fragments of pathological TDP-43 in amyotrophic lateral sclerosis and frontotemporal lobar degeneration. Biochem. Biophys. Res. Commun. 417, 116–121 (2012).
Tarutani, A. & Hasegawa, M. in Experimental Models of Parkinson’s Disease (ed. Imai, Y.) 17–25 (Springer, 2021).
Kametani, F. et al. Comparison of common and disease-specific post-translational modifications of pathological tau associated with a wide range of tauopathies. Front. Neurosci. 14, 581936 (2020).
Zivanov, J., Nakane, T. & Scheres, S. H. W. A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ 6, 5–17 (2019).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
He, S. & Scheres, S. H. W. Helical reconstruction in RELION. J. Struct. Biol. 198, 163–176 (2017).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Scheres, S. H. W. Amyloid structure determination in RELION-3.1. Acta Crystallogr. D Struct. Biol. 76, 94–101 (2020).
Zivanov, J., Nakane, T. & Scheres, S. H. W. Estimation of high-order aberrations and anisotropic magnification from cryo-EM data sets in RELION-3.1. IUCrJ 7, 253–267 (2020).
Chen, S. et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryo-microscopy. Ultramicroscopy 135, 24–35 (2013).
Casañal, A., Lohkamp, B. & Emsley, P. Current developments in Coot for macromolecular model building of electron cryo-microscopy and erystallographic data. Protein Sci. 29, 1055–1064 (2020).
Croll, T. I. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. D Struct. Biol. 74, 519–530 (2018).
Brown, A. et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr. D Biol. Crystallogr. 71, 136–153 (2015).
Yamashita, K., Palmer, C. M., Burnley, T. & Murshudov, G. N. Cryo-EM single -particle structure refinement and map calculation using Servalcat. Acta Crystallogr. D Struct. Biol. 77, 1281–1291 (2021).
Williams, C. J. et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Acknowledgements
We thank the individuals and their families for donating the brain tissue; K. Yamada and Y. Itoh for the clinical information; A. Akagi for dissecting the brain tissue; T. Arai and H. Akiyama for helpful comments on the clinical history and neuropathology; R. Otani and M. Takase for technical support with immunohistochemistry; K. Yamashita and G. N. Murshudov for help with Servalcat; staff at the MRC Laboratory of Molecular Biology Electron Microscopy Facility for access to and support with EM; staff at the MRC Laboratory of Molecular Biology Scientific Computing Facility for access to and support with computing; and M. Goedert, S. H. W. Scheres, S.W. Davies, S. Tetter, T. S. Behr and R. R. Chen for discussions. D.A. is a Fellow at Darwin College, University of Cambridge. This work was supported by the Medical Research Council (MRC) grant MC_UP_1201/25 and Alzheimer’s Research UK award ARUK-RS2019-001 to B.R.-F.; the Japan Agency for Medical Research and Development (AMED) grants JP20ek0109392 and JP20ek0109391 to M.Y. and JP20dm0207072 to M.H; and the Japan Science and Technology Agency (JST) Core Research for Evolutional Science and Technology (CREST) grant JPMJCR18H3 to M.H.
Author information
Authors and Affiliations
Contributions
M.Y. identified the individuals and performed the neuropathological examinations. M.A. performed the genetic analysis. M.H. extracted the TDP-43 filaments and performed immunohistochemistry, immunoblotting and immuno-EM. F.K. performed the mass spectrometry. D.A. performed cryo-EM. D.A. and B.R.-F. analysed the cryo-EM data. D.A., B.R.-F. and A.G.M. built and analysed the atomic model. B.R.-F. supervised the study. B.R.-F., M.Y. and M.H. are the senior authors. All authors contributed to writing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks James Shorter, Henning Stahlberg and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 TDP-43 pathology of ALS with FTLD continued.
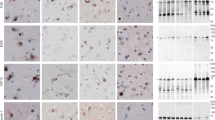
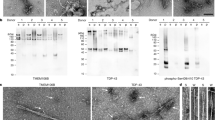
a, Staining of TDP-43 (brown) in the frontal and motor cortices and spinal cords of individuals 1 and 2 with anti-TDP-43 (amino acids 203–209) antibody. Nuclei were counterstained in blue. Scale bars, 50 μm. b, Staining of TDP-43 glial cytoplasmic inclusions (brown) in the motor cortices of individuals 1 and 2 with anti-phosphorylated S409 and S410 TDP-43 antibody. Nuclei were counterstained in blue. Scale bars, 50 μm. c, d, Immunoblots of the total homogenate (T), Sarkosyl-soluble fraction (S) and Sarkosyl-insoluble fraction (I) from the motor cortices of individuals 1 and 2 with anti-TDP-43 (amino acids 203–209) (c) and anti-phosphorylated S409 and S410 TDP-43 (d) antibodies. The original, uncropped blots are shown in Supplementary Fig. 1. e, Immuno-electron microscopy of the Sarkosyl-insoluble fraction from the frontal cortex of individual 2 using anti-phosphorylated S409 and S410 TDP-43 antibody. Scale bar, 100 nm. a–e, Similar results were obtained in at least three independent experiments.
Extended Data Fig. 2 Cryo-EM and helical reconstruction.
a, Representative cryo-EM images of TDP-43 filaments from the frontal and motor cortices of individual 1 and from the frontal cortex of individual 2. Scale bars, 100 nm. Similar results were obtained in at least three independent experiments. b, Fourier shell correlation (FSC) curves for two independently-refined cryo-EM half-maps. The FSC threshold of 0.143 is shown with red dashed lines. c, Local resolution estimates for the Cryo-EM 3D reconstructions. Scale bars, 10 Å. d, Cryo-EM density maps viewed along the helical axis. Scale bars, 10 Å.
Extended Data Fig. 3 Cryo-EM density map and atomic model comparisons.
a–c, Views of the cryo-EM density map of TDP-43 filaments from the frontal cortex of individual 1 and the corresponding atomic model showing representative densities for amino acid side chains (a), peptide group oxygen atoms (b) and ordered solvent molecules (red arrows) (c). d, Fourier shell correlation (FSC) curves for the refined atomic model against the cryo-EM density map (black); for the atomic model shaken and refined using the first half-map against the first half-map (magenta); and for the same atomic model against the second half-map (green). The FSC threshold of 0.5 is shown with a red dashed line.
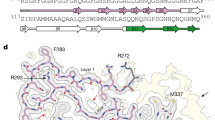
Extended Data Fig. 4 Double-spiral fold of TDP-43 filaments from ALS with FTLD.
a, Schematic view of the double-spiral fold. The two hydrophobic clusters in the hydrophobic nucleus, composed of the side chains of M307, M311, F313, M322, M323, A326, A329 and L330 (cluster 1); and A328, W334, M336 and L340 (cluster 2), are numbered. b, Hydrophobicity of the double-spiral fold from most hydrophilic (cyan) to most hydrophobic (yellow). c, Secondary structure of the double-spiral fold, depicted as five successive rungs. The glycine-rich (G282-G310, magenta), hydrophobic (M311-S342, white) and Q/N-rich (Q343-Q360, green) regions are highlighted. d, Views of the double-spiral fold, depicted as three successive rungs, showing hydrogen bonds (blue dashed lines) between buried polar side chains and main chain peptide groups. e, Superposition of residues N319-A326 of the double-spiral fold (cyan), depicted as two successive rungs, with residues G88-M95 and G107-A114 of the β-helix domain of glutamate synthase (magenta, PDB ID 1EA0). f, Unmasked cryo-EM 3D reconstructions of TDP-43 filaments from the frontal and motor cortices of individual 1 and from the frontal cortex of individual 2, shown as central slices perpendicular to the helical axis. Additional density within the prominent groove on the filament surface formed by the main chain of G282–Q286 and the side chain of Q286 is indicated with a magenta arrow. Additional densities adjacent to flat strips on the surface of the glycine-rich spiral branch between R293 and A315 are indicated with yellow arrows. Additional less well-resolved density projecting from a polar patch formed by the side chains of N352, Q354 and Q356 on the surface of the Q/N-rich spiral branch is indicated with a cyan arrow. Scale bars, 25 Å.
Extended Data Fig. 5 Comparison of the double-spiral fold with recombinant TDP-43 structures.
a, Amino acid sequence alignment of the secondary structure elements of the double-spiral fold and recombinant TDP-43 structures with the TDP-43 LC domain. PDB IDs are given for the recombinant TDP-43 structures. The glycine-rich (G282-G310, magenta), hydrophobic (M311-S342, white) and Q/N-rich (Q343-Q360, green) regions are highlighted on the double-spiral fold secondary structure elements. b, Secondary structure of the double-spiral fold and recombinant TDP-43. PDB IDs are given for the recombinant TDP-43 structures. The glycine-rich (G282-G310, magenta), hydrophobic (M311-S342, white) and Q/N-rich (Q343-Q360, green) regions are highlighted.
Extended Data Fig. 6 Heat stability of TDP-43 filaments from ALS with FTLD.
Representative cryo-EM images of TDP-43 filaments from the frontal cortex of individual 1 with and without heating at 65 °C for 10 min. Scale bars, 100 nm. Similar results were obtained in at least three independent experiments.
Supplementary information
Supplementary Figure
Uncropped images of immunoblots presented in this study.
Rights and permissions
About this article
Cite this article
Arseni, D., Hasegawa, M., Murzin, A.G. et al. Structure of pathological TDP-43 filaments from ALS with FTLD. Nature 601, 139–143 (2022). https://doi.org/10.1038/s41586-021-04199-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-04199-3
This article is cited by
-
Misfolded protein oligomers: mechanisms of formation, cytotoxic effects, and pharmacological approaches against protein misfolding diseases
Molecular Neurodegeneration (2024)
-
Synthetic β-sheets mimicking fibrillar and oligomeric structures for evaluation of spectral X-ray scattering technique for biomarker quantification
Cell & Bioscience (2024)
-
Nuclear-import receptors as gatekeepers of pathological phase transitions in ALS/FTD
Molecular Neurodegeneration (2024)
-
Neuropathogenesis-on-chips for neurodegenerative diseases
Nature Communications (2024)
-
RNA modulates hnRNPA1A amyloid formation mediated by biomolecular condensates
Nature Chemistry (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.