Abstract
The human microbiome encodes a large repertoire of biochemical enzymes and pathways, most of which remain uncharacterized. Here, using a metagenomics-based search strategy, we discovered that bacterial members of the human gut and oral microbiome encode enzymes that selectively phosphorylate a clinically used antidiabetic drug, acarbose1,2, resulting in its inactivation. Acarbose is an inhibitor of both human and bacterial α-glucosidases3, limiting the ability of the target organism to metabolize complex carbohydrates. Using biochemical assays, X-ray crystallography and metagenomic analyses, we show that microbiome-derived acarbose kinases are specific for acarbose, provide their harbouring organism with a protective advantage against the activity of acarbose, and are widespread in the microbiomes of western and non-western human populations. These results provide an example of widespread microbiome resistance to a non-antibiotic drug, and suggest that acarbose resistance has disseminated in the human microbiome as a defensive strategy against a potential endogenous producer of a closely related molecule.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Associated data are provided in Extended Data Figs. 1–9 and Supplementary Tables 1–10. Structures of seleno-methionine Mak1, Mak1 bound to AMP-PNP and acarbose and AcbK bound to AMP-PMP and acarbose are available in the Protein Data Bank under accession codes 6WB4, 6WB5 and 6WB7, respectively. Publicly available datasets used in the metagenomic analyses performed here were obtained as follows: HMP-1-1 (43021), Chinese (PRJNA422434), MetaHIT (PRJEB1220), Fijicomp (PRJNA217052), HMP-1-2 (PRJNA275349, PRJNA48479), human clinical trial with acarbose (PRJEB14155). The following datasets were used in metatranscriptomic analyses: PRJNA354235, PRJNA389280, PRJNA398089, PRJEB4673, PRJNA221620, https://www.mg-rast.org/linkin.cgi?project=mgp5148, PRJNA383868.
Code availability
MetaBGC (v.1.3.3, https://github.com/donia-lab/MetaBGC) was used to discover AcbK homologues in unassembled metagenomic sequencing data, following the details and parameters described in the Methods.
References
Chiasson, J. L. et al. Acarbose for prevention of type 2 diabetes mellitus: the STOP-NIDDM randomised trial. Lancet 359, 2072–2077 (2002).
Wehmeier, U. F. & Piepersberg, W. Biotechnology and molecular biology of the alpha-glucosidase inhibitor acarbose. Appl. Microbiol. Biotechnol. 63, 613–625 (2004).
Yoon, S.-H. & Robyt, J. F. Study of the inhibition of four alpha amylases by acarbose and its 4IV-α-maltohexaosyl and 4IV-α-maltododecaosyl analogues. Carbohydr. Res. 338, 1969–1980 (2003).
Maurice, C. F., Haiser, H. J. & Turnbaugh, P. J. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell 152, 39–50 (2013).
Wu, H. et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat. Med. 23, 850–858 (2017).
Maier, L. et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 555, 623–628 (2018).
Whang, A., Nagpal, R. & Yadav, H. Bi-directional drug-microbiome interactions of anti-diabetics. eBioMedicine 39, 591–602 (2019).
Le Bastard, Q. et al. Systematic review: human gut dysbiosis induced by non-antibiotic prescription medications. Aliment. Pharmacol. Ther. 47, 332–345 (2018).
Maruhama, Y. et al. Effects of a glucoside-hydrolase inhibitor (Bay g 5421) on serum lipids, lipoproteins and bile acids, fecal fat and bacterial flora, and intestinal gas production in hyperlipidemic patients. Tohoku J. Exp. Med. 132, 453–462 (1980).
Su, B. et al. Acarbose treatment affects the serum levels of inflammatory cytokines and the gut content of bifidobacteria in Chinese patients with type 2 diabetes mellitus. J. Diabetes 7, 729–739 (2015).
Zhang, X. et al. Effects of acarbose on the gut microbiota of prediabetic patients: a randomized, double-blind, controlled crossover trial. Diabetes Ther. 8, 293–307 (2017).
Santilli, A. D., Dawson, E. M., Whitehead, K. J. & Whitehead, D. C. Nonmicrobicidal small molecule inhibition of polysaccharide metabolism in human gut microbes: a potential therapeutic avenue. ACS Chem. Biol. 13, 1165–1172 (2018).
Baxter, N. T., Lesniak, N. A., Sinani, H., Schloss, P. D. & Koropatkin, N. M. The glucoamylase inhibitor acarbose has a diet-dependent and reversible effect on the murine gut microbiome. mSphere 4, https://doi.org/10.1128/mSphere.00528-18 (2019).
Zhang, M. et al. Effects of metformin, acarbose, and sitagliptin monotherapy on gut microbiota in Zucker diabetic fatty rats. BMJ Open Diabetes Res. Care 7, e000717 (2019).
Ahr, H. J. et al. Pharmacokinetics of acarbose. Part I: absorption, concentration in plasma, metabolism and excretion after single administration of [14C]acarbose to rats, dogs and man. Arzneimittelforschung 39, 1254–1260 (1989).
Zhao, L. et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science 359, 1151–1156 (2018).
Wehmeier, U. F. The biosynthesis and metabolism of acarbose in Actinoplanes sp. SE 50/110: a progress report. Biocatal. Biotransform. 21, 279–284 (2003).
Schmidt, D. D. et al. Alpha-glucosidase inhibitors. New complex oligosaccharides of microbial origin. Naturwissenschaften 64, 535–536 (1977).
Drepper, A. & Pape, H. Acarbose 7-phosphotransferase from Actinoplanes sp.: purification, properties, and possible physiological function. J. Antibiot. 49, 664–668 (1996).
Goeke, K., Drepper, A. & Pape, H. Formation of acarbose phosphate by a cell-free extract from the acarbose producer Actinoplanes sp. J. Antibiot. 49, 661–663 (1996).
Human Microbiome Project, C. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
Qin, J. et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60 (2012).
Nielsen, H. B. et al. Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat. Biotechnol. 32, 822–828 (2014).
Brito, I. L. et al. Mobile genes in the human microbiome are structured from global to individual scales. Nature 535, 435–439 (2016).
Lloyd-Price, J. et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature 550, 61–66 (2017).
Sugimoto, Y. et al. A metagenomic strategy for harnessing the chemical repertoire of the human microbiome. Science 366, eaax9176 (2019).
Rockser, Y. & Wehmeier, U. F. The gac-gene cluster for the production of acarbose from Streptomyces glaucescens GLA.O: identification, isolation and characterization. J. Biotechnol. 140, 114–123 (2009).
Guo, X. et al. Draft genome sequence of Streptomyces coelicoflavus ZG0656 reveals the putative biosynthetic gene cluster of acarviostatin family alpha-amylase inhibitors. Lett. Appl. Microbiol. 55, 162–169 (2012).
Parducci, R. E., Cabrera, R., Baez, M. & Guixe, V. Evidence for a catalytic Mg2+ ion and effect of phosphate on the activity of Escherichia coli phosphofructokinase-2: regulatory properties of a ribokinase family member. Biochemistry 45, 9291–9299 (2006).
Miller, B. G. & Raines, R. T. Identifying latent enzyme activities: substrate ambiguity within modern bacterial sugar kinases. Biochemistry 43, 6387–6392 (2004).
Fong, D. H. & Berghuis, A. M. Substrate promiscuity of an aminoglycoside antibiotic resistance enzyme via target mimicry. EMBO J. 21, 2323–2331 (2002).
McAuley, M., Huang, M. & Timson, D. J. Dynamic origins of substrate promiscuity in bacterial galactokinases. Carbohydr. Res. 486, 107839 (2019).
Sigrell, J. A., Cameron, A. D., Jones, T. A. & Mowbray, S. L. Structure of Escherichia coli ribokinase in complex with ribose and dinucleotide determined to 1.8 å resolution: insights into a new family of kinase structures. Structure 6, 183–193 (1998).
Yeung, M. K. & Kozelsky, C. S. Transformation of Actinomyces spp. by a gram-negative broad-host-range plasmid. J. Bacteriol. 176, 4173–4176 (1994).
Flint, H. J., Scott, K. P., Duncan, S. H., Louis, P. & Forano, E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 3, 289–306 (2012).
Patnode, M. L. et al. Interspecies competition impacts targeted manipulation of human gut bacteria by fiber-derived glycans. Cell 179, 59–73 (2019).
Rakoff-Nahoum, S., Coyne, M. J. & Comstock, L. E. An ecological network of polysaccharide utilization among human intestinal symbionts. Curr. Biol. 24, 40–49 (2014).
Leimena, M. M. et al. A comprehensive metatranscriptome analysis pipeline and its validation using human small intestine microbiota datasets. BMC Genom. 14, 530 (2013).
Goodrich, J. K. et al. Genetic determinants of the gut microbiome in UK twins. Cell Host Microbe 19, 731–743 (2016).
Villmones, H. C. et al. Species level description of the human ileal bacterial microbiota. Sci. Rep. 8, 4736 (2018).
Fung, T. C. et al. Intestinal serotonin and fluoxetine exposure modulate bacterial colonization in the gut. Nat. Microbiol. 4, 2064–2073 (2019).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016).
Abu-Ali, G. S. et al. Metatranscriptome of human faecal microbial communities in a cohort of adult men. Nat. Microbiol. 3, 356–366 (2018).
Schirmer, M. et al. Dynamics of metatranscription in the inflammatory bowel disease gut microbiome. Nat. Microbiol. 3, 337–346 (2018).
Lloyd-Price, J. et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569, 655–662 (2019).
Peterson, S. N. et al. Functional expression of dental plaque microbiota. Front. Cell Infect. Microbiol. 4, 108 (2014).
Benitez-Paez, A., Belda-Ferre, P., Simon-Soro, A. & Mira, A. Microbiota diversity and gene expression dynamics in human oral biofilms. BMC Genom. 15, 311 (2014).
Jorth, P. et al. Metatranscriptomics of the human oral microbiome during health and disease. mBio 5, e01012-14 (2014).
Szafranski, S. P. et al. Functional biomarkers for chronic periodontitis and insights into the roles of Prevotella nigrescens and Fusobacterium nucleatum; a metatranscriptome analysis. NPJ Biofilms Microbiomes 1, 15017 (2015).
Schmieder, R. & Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27, 863–864 (2011).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Chen, I. A. et al. IMG/M v.5.0: an integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res. 47, D666–D677 (2019).
Kabsch, W. Xds. Acta Crystallogr. D 66, 125–132 (2010).
Evans, P. R. & Murshudov, G. N. How good are my data and what is the resolution? Acta Crystallogr. D 69, 1204–1214 (2013).
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D 67, 235–242 (2011).
Sheldrick, G. M. A short history of SHELX. Acta Crystallogr. A 64, 112–122 (2008).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Kearse, M. et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649 (2012).
Acknowledgements
We thank the NSLS-2 AMX and FMX beamline staff for their assistance with data collection and the crystallography core facility at the Department of Molecular Biology, Princeton University. We thank M. Cahn and M. Elmassry for assistance with metagenomic data analysis; S. Chatterjee for general assistance; and the rest of the Donia laboratory for discussions. Funding for this project was provided by an NIH Director’s New Innovator Award (1DP2AI124441) and the Pew Biomedical Scholars Program to M.S.D.; an NIH grant (1R01GM110161), a Burroughs Wellcome Foundation Grant (1013579) and an award from The Vallee Foundation to A.K. J.B. is funded by a training grant from the National Institute of General Medicine Sciences (NIGMS) (T32GM007388) and L.Z. is a CIFAR fellow. This research used the AMX and FMX beamlines of the National Synchrotron Light Source II, a United States Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Brookhaven National Laboratory under contract no. DE-SC0012704. The Life Science Biomedical Technology Research resource, which supports AMX and FMX, is primarily supported by the NIH (NIGMS) through a Biomedical Technology Research Resource P41 grant (P41GM111244), and by the DOE Office of Biological and Environmental Research (KP1605010).
Author information
Authors and Affiliations
Contributions
M.S.D. and J.B. conceived the project. J.B. and M.E. performed biochemical experiments and analysed the resulting data, with input from M.S.D. and A.K.; P.D.J. collected and analysed X-ray crystallography data, with input from J.B., M.E., M.S.D. and A.K.; and J.B. performed microbiological experiments and analysed the resulting data. A.B., J.B. and M.S.D. performed metagenomic and metatranscriptomic data analyses. G.W. and L.Z. performed human clinical data analysis. J.B., P.D.J. and M.S.D. wrote the paper, with input from all of the other authors.
Corresponding author
Ethics declarations
Competing interests
M.S.D. is a member of the scientific advisory board of Deepbiome Therapeutics, and L.Z. is a co-founder of Notitia Biotechnologies.
Additional information
Peer review information Nature thanks Sofia Forslund and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Identification of AcbK homologues using MetaBGC.
a. Number of true positive (TP) reads identified in synthetic metagenomic dataset number 2 for all spHMMs created by MetaBGC-Build. b. Number of false positive (FP) reads identified in synthetic metagenomic dataset number 2 for all spHMMs created by MetaBGC-Build. c. F1 scores of all spHMMs created by MetaBGC-Build. Only spHMMs with F1 scores ≥ 0.5 were used in MetaBGC-Identify runs with real metagenomic data. d. spHMM logos for spHMMs included in MetaBGC-Identify. Please see Supplementary Table 2 for spHMM cutoffs used for all intervals.
Extended Data Fig. 2 Phylogenetic analysis of Mcks/Maks and prevalence of selected maks and pmaks in metagenomic samples of various human cohorts.
a. A maximum likelihood phylogenetic tree for all AcbK homologues discovered in this study (Mcks). The tree was constructed using MEGA7, with bootstrap values of more than 50% out of 1000 replicates displayed at the branch points (see Methods). The tree includes previously characterized acarbose kinases from soil-derived Actinobacteria (red label, AcbK, GacK, and ScatK), the canonical phosphofructokinase from E. coli (PfkB), Mcks that were experimentally verified to have (green label, designated as Maks) or lack (blue label) an acarbose-O6A-kinase activity, and ones that have not been experimentally tested (black label). pMaks indicate putative Maks in the AcbK clade. b. Prevalence of selected maks and pmaks expressed as the percentage of positive individuals out of the total, across different cohorts and sampling sites and subsites (brown: gut; teal: oral). The total number of individuals in each cohort is indicated above their corresponding bar chart. Prevalence of all mcks in all cohorts can be found in Supplementary Table 2.
Extended Data Fig. 3 Purification and biochemical characterization of a subset of Mcks/Maks.
a. Fast Protein Liquid Chromatography (FPLC) chromatograms of AcbK (a) and Mck1/Mak1 (b) purification by size exclusion chromatography, monitored at the indicated absorbance wavelengths. Coomassie stained SDS-PAGE of the collected fraction is shown on the right side of each chromatogram. c. Coomassie stained SDS-PAGE of the collected fractions for the eight additional Mcks that were tested. All Mcks were purified in the same manner as AcbK and Mck1/Mak1 (see Methods). The data shown in a, b are representative examples from among at least three different times the same experiment was performed and produced similar results. The data shown in in c is a representative example from among at least two different times the same experiment was performed and produced similar results. d. Representative Extracted Ion Chromatograms (EICs) for acarbose (m/z = 646.4, [M+H]+) from the products of a reaction of Mck1/Mak1 (red) or a no-protein control (blue) with acarbose. e. Representative EICs for acarbose-O6A-phosphate (m/z = 726.4, [M+H]+) from the products of a reaction of Mck1/Mak1 (red) or a no-protein control (blue) with acarbose (the two peaks have identical MS, HRMS, and HRMS/MS and are likely isomers of the same molecule or charge variants that elute differently, as seen with AcbK, see Extended Data Fig. 5). This product is often referred to as acarbose-7-phosphate in the literature, but for consistency, we will refer to all acarbose positions based on their displayed numbering in Fig. 1a, which matches previous PDB depositions (PDB Ligand Code: ACR). Note the complete conversion of acarbose to acarbose-O6A-phosphate. See Methods for the complete experimental details.
Extended Data Fig. 4 Abundances of all metagenomic read bins discovered by MetaBGC.
Heatmap showing the abundance of all identified bins in the five analysed cohorts as calculated by MetaBGC, and following the colour codes to the right. See Supplementary Table 2 for the complete results of this analysis and Supplementary Table 1 for corresponding Mck/Mak identifiers for each bin. Samples that had no reads mapped to any bin were excluded from the heatmap. Hierarchical clustering of the samples and bins was performed using UPGMA (unweighted pair group method with arithmetic mean) in pheatmap in R.
Extended Data Fig. 5 HPLC-HR–MS and HPLC-HR–MS/MS analyses of acarbose-O6A-phosphate produced by AcbK and Mak1.
Extracted Ion Chromatograms (EICs) of acarbose-O6A-phosphate produced by AcbK (a), Mak1 (d), and a co-injection of the two (f), indicating that their products are identical. EIC is displayed for m/z = 726.2-726.3 [M+H]+ in all cases. b, c, e, g, HR-MS of the two acarbose-O6A-phosphate peaks produced by either AcbK (b, c) or Mak1 (e, g), following the numbering scheme on the individual EIC peaks. h, i, identical HR-MS/MS fragmentation patterns of the two EIC peaks of acarbose-O6A-phosphate from the co-injection analysis in f. The structure of acarbose-O6A-phosphate is shown on the right, along with its predicted fragments and their calculated mass to charge ratios that match observed ones.
Extended Data Fig. 6 Additional biochemical characterization of Mak1 and AcbK.
a. Phosphorylation rates (relative to the acarbose phosphorylation rate, which is set as 100%) at which Mak1 and AcbK phosphorylate a diverse panel of carbohydrates and aminoglycosides with structural similarities to acarbose. No phosphorylation was detected for all but one (validamycin) of the substrates under the same experimental conditions (see Methods). Experiments were done in duplicates with the average value used for rate comparisons; raw data is available in Supplementary Table 3. b. Michaelis-Menten saturation curves for AcbK (grey) and Mak1 (blue) performed at 1 μm enzyme concentration. Km and kcat values are indicated in their respective colours and individual kobs measurement replicates are shown on the graph for both enzymes. Raw data is available in Supplementary Table 3. c. kobs of both AcbK (grey) and Mak1 (blue) across different temperatures from 25-40 °C in 3 °C steps. The difference in kobs between the two enzymes can be seen across different temperatures. d, e. Hill plot (logarithm of kobs on the the y axis and logarithm of protein concentration on the x axis) of Mak1 (d) and AcbK (e). The Hill coefficient (slope) is greater than 1 for Mak1 (d, n=1.71 ± 0.06, n=1 is shown in grey dashed line for reference) but not for AcbK (e, n=1.13 ± 0.02), suggesting that only Mak1 is a cooperative enzyme. f. Relative change in kobs (y axis) for the single mutants D160A and D247A as well as the double mutant (D160A, H162A) of Mak1 as compared to wild type protein (N= 2). g. Extracted Ion Chromatograms (EICs) for acarbose (left, m/z = 646.4, [M+H]+) and acarbose-O6A-phosphate (right, m/z = 726.4, [M+H]+), showing that the addition of EDTA (blue traces) abolishes the activity of Mak1, while the addition of excess MgCl2 (red traces) restores it.
Extended Data Fig. 7 Additional structural details of AcbK.
a. AcbK forms homodimers with extensive interactions between the two monomers. These interactions include multiple β strands in the β-clasp domain, most notably the β3 of one monomer and the β8 of the other (magenta highlighted box, with β strands from each monomer shaded differently for clarity). A molecular surface view is shown on the right with one AcbK monomer coloured green and the other coloured blue, highlighting the extensive surface area (1,298 Å2) involved in forming the dimer via the β-clasp domains. b. Zoomed in view of the AcbK substrate binding pocket where an extensive network of hydrogen bonds (from residues Asp16, Asn99, Ser109, and Asp248) form with all the hydroxyl groups in the acarbose A ring and hold it in place. Distances are shown for each of the hydrogen bonds mentioned above. c. Zoomed in view of the AcbK active site with important residues shown (Asp162, His164, Asp248), all involved in priming the O6A hydroxyl of acarbose for nucleophilic attack and in facilitating the transfer of the phosphate from an ATP (AMP-PNP shown) to the O6A hydroxyl of acarbose. Distance shown is from the O6A hydroxyl to the γ-phosphate. d. Full-length amino acid sequence alignment of experimentally tested Mcks/Maks, as well as AcbK and PfkB. The bars on the top of the alignment denote the average pairwise percent identity at each residue. e. Selected segments of the amino acid sequence alignment of experimentally tested Mcks/Maks, as well as AcbK and PfkB. Blue colours highlight amino acid residues that are deemed important for hydrogen bonding with the A ring of acarbose, yellow colours highlight those involved in the transfer of the phosphate group to acarbose and green indicates a residue involved in both processes. Grey colours indicate other conserved residues in the alignment. The bars on the top of the alignment denote the average pairwise percent identity at each residue.
Extended Data Fig. 8 Additional functional and genetic analyses of maks and end.
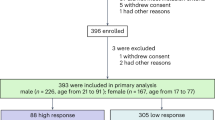
a. Normalized bacterial growth at 48 h, measured as the optical density at 600 nm and presented as a percentage of the untreated control. Different bars indicate treatments with varying acarbose concentrations for each of the tested strains: A. viscosus expressing mak1 (blue) and A. viscosus harbouring an empty-vector control (grey). mak1 expression resulted in a statistically significant rescue of the carbohydrate-dependent growth inhibitory activity of acarbose at several concentrations. Error bars represent the standard error of the mean among four replicates and “*” denotes a p-value < 0.01 (using a two-sample t-test assuming unequal variances): 0.5 µM, p=3.33x10-5; 5 µM, p=2.17x10−5; 10 µM, p=2.37x10−8; 50 µM, p=2.96x10−5; and 100 µM, p=5.94x10−3. See Supplementary Table 6 for the raw experimental data. b, c. HbA1c (b) and fasting blood glucose (c) levels in mak-negative (N=8, blue) and mak-positive (N=8, red) Type 2 Diabetes patients treated with acarbose. The bar graphs show mean ± SD. Repeated measures analysis of variance (ANOVA) with Tukey’s post hoc test was used for comparing different time points in each group. Compact letter displays were used to indicate the statistical differences (P < 0.05) between different time points. The difference between groups at each post-treatment time point was evaluated by an ANCOVA model controlling for the baseline measurements. * P < 0.05. We observed statistically significant reductions in both HbA1c and fasting blood glucose levels only in mak-negative patients. In addition, at day 84, mak-negative patients showed significantly larger reduction of HbA1c (PANCOVA = 0.011) and fasting blood glucose levels (PANCOVA = 0.024) from baseline than mak-positive patients. d. Genetic context of all maks and pmaks, as well as acbK, gacK and scatK, following the colour key to the right. e. A simplified biosynthetic scheme for acarbose by gac, with gac proteins involved at each step shown above the arrows. Homologous end proteins are shown below the arrow, accounting for most of the core biosynthetic steps needed for acarbose production.
Extended Data Fig. 9 Distribution of mak1 and end variants in the human microbiome.
a. Percentage of samples across different cohorts that are positive for either mak1 (blue) or end (green). mak1 was considered “present” in a given sample if any reads mapped to it, and end was considered “present” in a given sample if it had a breadth coverage of >25% of its length. SP stands for supragingival plaque. See Supplementary Table 8 for detailed results of this analysis. b. Genetic context of all five end variants discovered, following the colour code on the right. Note that most variants exist in close proximity to a transposase gene, suggesting a role in their mobility. c. Relative RPKM (y axis, defined as the RPKM of the gene of interest divided by the RPKM of the entire end cluster) of each end gene (x axis). Ten examples of human metagenomic samples (supragingival plaque, HMP) are shown in the top graph, where the depth of coverage is uniform across all end genes. Ten examples of human metagenomic samples (supragingival plaque, HMP) are shown in the bottom graph, where a clear spike in the depth of coverage can be observed around the endM/mak1 gene, indicating the presence of at least two different end genetic variants in these samples. Next to each graph is a representative example of metagenomic reads from supragingival plaque samples mapped to the end BGC. See Supplementary Table 10 for detailed results of this analysis. d. A heatmap showing the abundance (in RPKM) of all end genes in supragingival plaque samples of the HMP cohorts (HMP-1-1 and HMP-1-2). Different samples harbour different genetic contexts (or variants) of the end BGC: 14-gene, 8-gene, 4-gene, 2-gene, or stand-alone mak1/endM. Samples are sorted according to their classification into one of these five genetic variants listed above. e. Metagenomic reads from five different supragingival plaque samples mapped to the end BGC (see Methods). Each of the five samples illustrates an example for one of the unique genetic variants described above. f. A Pie chart showing the distribution of the five genetic variants amongst HMP participants (supragingival plaque samples). If participants had multiple visits and different variants across visits, they were classified into end (multiple variants) (see Supplementary Table 10).
Supplementary information
Supplementary Figure 1
Raw unedited images for gels shown in Extended Data Figure 3. a, Raw unedited gel image that corresponds to the cropped gel image shown in Extended Data Figure 3a. The ladder bands are labelled on the y-axis and the x-axis labels indicate sample names. Samples labelled as NA belong to an experiment unrelated to this manuscript. b, Raw unedited gel image that corresponds to the cropped gel image shown in Extended Data Figure 3b. The ladder bands are labelled on the y-axis and the x-axis labels indicate sample names. c, Raw unedited gel image that corresponds to the cropped gel image shown in Extended Data Figure 3c. The ladder bands are labelled on the y-axis and the x-axis labels indicate sample.
Supplementary Table 1
Additional information about AcbK homologues identified from all computational searches.
Supplementary Table 2
Details and results of MetaBGC analyses.
Supplementary Table 3
Raw enzymatic assay data for Maks/Mcks characterized in this study.
Supplementary Table 4
Raw data for the amylase inhibition assay with acarbose and acarbose-O6A-phosphate.
Supplementary Table 5
AcbK and Mak1 crystal structure refinement statistics.
Supplementary Table 6
Raw data for the A. viscosus growth rescue experiment.
Supplementary Table 7
Quantification of mak and pmak genes in faecal metagenomic datasets from a recent acarbose human clinical trial16.
Supplementary Table 8
Quantification of mak1 and end across all samples and cohorts.
Supplementary Table 9
Comparison between the end and gac BGCs.
Supplementary Table 10
Analysis of end genetic variants in supragingival plaque samples of HMP participants.
Rights and permissions
About this article
Cite this article
Balaich, J., Estrella, M., Wu, G. et al. The human microbiome encodes resistance to the antidiabetic drug acarbose. Nature 600, 110–115 (2021). https://doi.org/10.1038/s41586-021-04091-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-04091-0
This article is cited by
-
Roles of the gut microbiome in weight management
Nature Reviews Microbiology (2023)
-
Targeting the human gut microbiome with small-molecule inhibitors
Nature Reviews Chemistry (2023)
-
Inactivation of the antidiabetic drug acarbose by human intestinal microbial-mediated degradation
Nature Metabolism (2023)
-
Gut bacteria thwart the blood sugar-lowering effect of acarbose
Nature Metabolism (2023)
-
Gut microbiota: a potential target for improved cancer therapy
Journal of Cancer Research and Clinical Oncology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.