Abstract
Bacterial chemotaxis, the directed movement of cells along gradients of chemoattractants, is among the best-characterized subjects in molecular biology1,2,3,4,5,6,7,8,9,10, but much less is known about its physiological roles11. It is commonly seen as a starvation response when nutrients run out, or as an escape response from harmful situations12,13,14,15,16. Here we identify an alternative role of chemotaxis by systematically examining the spatiotemporal dynamics of Escherichia coli in soft agar12,17,18. Chemotaxis in nutrient-replete conditions promotes the expansion of bacterial populations into unoccupied territories well before nutrients run out in the current environment. Low levels of chemoattractants act as aroma-like cues in this process, establishing the direction and enhancing the speed of population movement along the self-generated attractant gradients. This process of navigated range expansion spreads faster and yields larger population gains than unguided expansion following the canonical Fisher–Kolmogorov dynamics19,20 and is therefore a general strategy to promote population growth in spatially extended, nutrient-replete environments.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Major experimental data that support this study are provided in this manuscript or available via figshare repositories: https://figshare.com/articles/Confocal_intensity_scans_expanding_bacteria/9639209 (confocal expansion data) and https://figshare.com/articles/Swimming_in_liquid_culture/9643001 (swimming observation data). Simulation data can be generated with the provided simulation code. Simulation parameters are provided in a separate file. The Supplementary Text provides additional details on strains, experimental methods and modelling.
Code availability
Custom-made code is available via GitHub for the analysis of swimming characteristics (https://github.com/jonascremer/swimming_analysis), the analysis of expanding populations using confocal microscopy (https://github.com/jonascremer/chemotaxis_imageanalysisexpansiondynamics), and the numerical simulations of the growth–expansion model (https://github.com/jonascremer/chemotaxis_simulation).
References
Berg, H. C. E. coli in Motion (Springer, 2004).
Alon, U., Surette, M. G., Barkai, N. & Leibler, S. Robustness in bacterial chemotaxis. Nature 397, 168–171 (1999).
Sourjik, V. & Berg, H. C. Functional interactions between receptors in bacterial chemotaxis. Nature 428, 437–441 (2004).
Bray, D. & Duke, T. Conformational spread: the propagation of allosteric states in large multiprotein complexes. Annu. Rev. Biophys. Biomol. Struct. 33, 53–73 (2004).
Korobkova, E., Emonet, T., Vilar, J. M. G., Shimizu, T. S. & Cluzel, P. From molecular noise to behavioural variability in a single bacterium. Nature 428, 574–578 (2004).
Tu, Y., Shimizu, T. S. & Berg, H. C. Modeling the chemotactic response of Escherichia coli to time-varying stimuli. Proc. Natl Acad. Sci. USA 105, 14855–14860 (2008).
Sourjik, V. & Wingreen, N. S. Responding to chemical gradients: bacterial chemotaxis. Curr. Opin. Cell Biol. 24, 262–268 (2012).
Tu, Y. Quantitative modeling of bacterial chemotaxis: signal amplification and accurate adaptation. Annu. Rev. Biophys. 42, 337–359 (2013).
Waite, A. J. et al. Non-genetic diversity modulates population performance. Mol. Syst. Biol. 12, 895 (2016).
Baym, M. et al. Spatiotemporal microbial evolution on antibiotic landscapes. Science 353, 1147–1151 (2016).
Hein, A. M., Carrara, F., Brumley, D. R., Stocker, R. & Levin, S. A. Natural search algorithms as a bridge between organisms, evolution, and ecology. Proc. Natl Acad. Sci. USA 113, 9413–9420 (2016).
Adler, J. Chemoreceptors in bacteria. Science 166, 1588–1597 (1969).
Maeda, K., Imae, Y., Shioi, J. I. & Oosawa, F. Effect of temperature on motility and chemotaxis of Escherichia coli. J. Bacteriol. 127, 1039–1046 (1976).
Amsler, C. D., Cho, M. & Matsumura, P. Multiple factors underlying the maximum motility of Escherichia coli as cultures enter post-exponential growth. J. Bacteriol. 175, 6238–6244 (1993).
Staropoli, J. F. & Alon, U. Computerized analysis of chemotaxis at different stages of bacterial growth. Biophys. J. 78, 513–519 (2000).
Paulick, A. et al. Mechanism of bidirectional thermotaxis in Escherichia coli. eLife 6, e26607 (2017).
Adler, J. Chemotaxis in bacteria. Science 153, 708–716 (1966).
Koster, D. A., Mayo, A., Bren, A. & Alon, U. Surface growth of a motile bacterial population resembles growth in a chemostat. J. Mol. Biol. 424, 180–191 (2012).
Skellam, J. G. Random dispersal in theoretical populations. Biometrika 38, 196–218 (1951).
Hastings, A. et al. The spatial spread of invasions: new developments in theory and evidence. Ecol. Lett. 8, 91–101 (2005).
Adler, J., Hazelbauer, G. L. & Dahl, M. M. Chemotaxis toward sugars in Escherichia coli. J. Bacteriol. 115, 824–847 (1973).
Saragosti, J. et al. Directional persistence of chemotactic bacteria in a traveling concentration wave. Proc. Natl Acad. Sci. USA 108, 16235–16240 (2011).
Wong-Ng, J., Melbinger, A., Celani, A. & Vergassola, M. The role of adaptation in bacterial speed races. PLOS Comput. Biol. 12, e1004974 (2016).
Fu, X. et al. Spatial self-organization resolves conflicts between individuality and collective migration. Nat. Commun. 9, 2177 (2018).
Hui, S. et al. Quantitative proteomic analysis reveals a simple strategy of global resource allocation in bacteria. Mol. Syst. Biol. 11, 784 (2015).
You, C. et al. Coordination of bacterial proteome with metabolism by cyclic AMP signalling. Nature 500, 301–306 (2013).
Barker, C. S., Prüss, B. M. & Matsumura, P. Increased motility of Escherichia coli by insertion sequence element integration into the regulatory region of the flhD operon. J. Bacteriol. 186, 7529–7537 (2004).
Liu, M. et al. Global transcriptional programs reveal a carbon source foraging strategy by Escherichia coli. J. Biol. Chem. 280, 15921–15927 (2005).
Zhao, K., Liu, M. & Burgess, R. R. Adaptation in bacterial flagellar and motility systems: from regulon members to ‘foraging’-like behavior in E. coli. Nucleic Acids Res. 35, 4441–4452 (2007).
Lauffenburger, D., Kennedy, C. R. & Aris, R. Traveling bands of chemotactic bacteria in the context of population growth. Bull. Math. Biol. 46, 19–40 (1984).
Adler, J. & Templeton, B. The effect of environmental conditions on the motility of Escherichia coli. J. Gen. Microbiol. 46, 175–184 (1967).
Fisher, R. The wave of advance of advantageous genes. Ann. Eugen. 7, 355–369 (1937).
Kolmogorov, A., Petrovsky, I. & Piscounov, N. Étude de l’équation de la diffusion avec croissance de la quantité de matière et son application à un problème biologique. Mosk. Univ. Bull. Math. 1, 37 (1937).
Keller, E. F. & Segel, L. A. Model for chemotaxis. J. Theor. Biol. 30, 225–234 (1971).
Keller, E. F. & Segel, L. A. Traveling bands of chemotactic bacteria: a theoretical analysis. J. Theor. Biol. 30, 235–248 (1971).
Novick-Cohen, A. & Segel, L. A. A gradually slowing travelling band of chemotactic bacteria. J. Math. Biol. 19, 125–132 (1984).
Budrene, E. O. & Berg, H. C. Dynamics of formation of symmetrical patterns by chemotactic bacteria. Nature 376, 49–53 (1995).
Brenner, M. P., Levitov, L. S. & Budrene, E. O. Physical mechanisms for chemotactic pattern formation by bacteria. Biophys. J. 74, 1677–1693 (1998).
Saragosti, J. et al. Mathematical description of bacterial traveling pulses. PLOS Comput. Biol. 6, e1000890 (2010).
Nossal, R. Growth and movement of rings of chemotactic bacteria. Exp. Cell Res. 75, 138–142 (1972).
Lapidus, I. R. & Schiller, R. A model for traveling bands of chemotactic bacteria. Biophys. J. 22, 1–13 (1978).
Tindall, M. J., Maini, P. K., Porter, S. L. & Armitage, J. P. Overview of mathematical approaches used to model bacterial chemotaxis II: bacterial populations. Bull. Math. Biol. 70, 1570–1607 (2008).
Wolfe, A. J. & Berg, H. C. Migration of bacteria in semisolid agar. Proc. Natl Acad. Sci. USA 86, 6973–6977 (1989).
Prüss, B. M., Nelms, J. M., Park, C. & Wolfe, A. J. Mutations in NADH:ubiquinone oxidoreductase of Escherichia coli affect growth on mixed amino acids. J. Bacteriol. 176, 2143–2150 (1994).
Yang, Y. et al. Relation between chemotaxis and consumption of amino acids in bacteria. Mol. Microbiol. 96, 1272–1282 (2015).
Hallatschek, O., Hersen, P., Ramanathan, S. & Nelson, D. R. Genetic drift at expanding frontiers promotes gene segregation. Proc. Natl Acad. Sci. USA 104, 19926–19930 (2007).
Seymour, J. R., Simó, R., Ahmed, T. & Stocker, R. Chemoattraction to dimethylsulfoniopropionate throughout the marine microbial food web. Science 329, 342–345 (2010).
Celani, A. & Vergassola, M. Bacterial strategies for chemotaxis response. Proc. Natl Acad. Sci. USA 107, 1391–1396 (2010).
Lyons, E., Freeling, M., Kustu, S. & Inwood, W. Using genomic sequencing for classical genetics in E. coli K12. PLoS ONE 6, e16717 (2011).
Soupene, E. et al. Physiological studies of Escherichia coli strain MG1655: growth defects and apparent cross-regulation of gene expression. J. Bacteriol. 185, 5611–5626 (2003).
Brown, S. D. & Jun, S. Complete genome sequence of Escherichia coli NCM3722. Genome Announc. 3, e008795 (2015).
Basan, M. et al. Overflow metabolism in Escherichia coli results from efficient proteome allocation. Nature 528, 99–104 (2015).
Parkinson, J. S. Complementation analysis and deletion mapping of Escherichia coli mutants defective in chemotaxis. J. Bacteriol. 135, 45–53 (1978).
Cayley, S., Record, M. T., Jr & Lewis, B. A. Accumulation of 3-(N-morpholino)propanesulfonate by osmotically stressed Escherichia coli K-12. J. Bacteriol. 171, 3597–3602 (1989).
Berg, H. C. & Turner, L. Chemotaxis of bacteria in glass capillary arrays. Escherichia coli, motility, microchannel plate, and light scattering. Biophys. J. 58, 919–930 (1990).
Masson, J.-B., Voisinne, G., Wong-Ng, J., Celani, A. & Vergassola, M. Noninvasive inference of the molecular chemotactic response using bacterial trajectories. Proc. Natl Acad. Sci. USA 109, 1802–1807 (2012).
Liu, W., Cremer, J., Li, D., Hwa, T. & Liu, C. An evolutionarily stable strategy to colonize spatially extended habitats. Nature https://doi.org/10.1038/s41586-019-1734-x (2019).
Shehata, T. E. & Marr, A. G. Effect of nutrient concentration on the growth of Escherichia coli. J. Bacteriol. 107, 210–216 (1971).
Schellenberg, G. D. & Furlong, C. E. Resolution of the multiplicity of the glutamate and aspartate transport systems of Escherichia coli. J. Biol. Chem. 252, 9055–9064 (1977).
Cremer, J. et al. Effect of flow and peristaltic mixing on bacterial growth in a gut-like channel. Proc. Natl Acad. Sci. USA 113, 11414–11419 (2016).
Shimizu, T. S., Tu, Y. & Berg, H. C. A modular gradient-sensing network for chemotaxis in Escherichia coli revealed by responses to time-varying stimuli. Mol. Syst. Biol. 6, 382 (2010).
Shoval, O. et al. Fold-change detection and scalar symmetry of sensory input fields. Proc. Natl Acad. Sci. USA 107, 15995–16000 (2010).
Lazova, M. D., Ahmed, T., Bellomo, D., Stocker, R. & Shimizu, T. S. Response rescaling in bacterial chemotaxis. Proc. Natl Acad. Sci. USA 108, 13870–13875 (2011).
Celani, A., Shimizu, T. S. & Vergassola, M. Molecular and functional aspects of bacterial chemotaxis. J. Stat. Phys. 144, 219–240 (2011).
Vaknin, A. & Berg, H. C. Physical responses of bacterial chemoreceptors. J. Mol. Biol. 366, 1416–1423 (2007).
Neumann, S., Grosse, K. & Sourjik, V. Chemotactic signaling via carbohydrate phosphotransferase systems in Escherichia coli. Proc. Natl Acad. Sci. USA 109, 12159–12164 (2012).
Guyer, J. E., Wheeler, D. & Warren, J. A. FiPy: partial differential equations with Python. Comput. Sci. Eng. 11, 6–15 (2009).
Dufour, Y. S., Gillet, S., Frankel, N. W., Weibel, D. B. & Emonet, T. Direct correlation between motile behavior and protein abundance in single cells. PLOS Comput. Biol. 12, e1005041 (2016).
Keegstra, J. M. et al. Phenotypic diversity and temporal variability in a bacterial signaling network revealed by single-cell FRET. eLife 6, e27455 (2017).
Frankel, N. W. et al. Adaptability of non-genetic diversity in bacterial chemotaxis. eLife 3, e03526 (2014).
Müller, M. J. I., Neugeboren, B. I., Nelson, D. R. & Murray, A. W. Genetic drift opposes mutualism during spatial population expansion. Proc. Natl Acad. Sci. USA 111, 1037–1042 (2014).
Möbius, W., Murray, A. W. & Nelson, D. R. How obstacles perturb population fronts and alter their genetic structure. PLOS Comput. Biol. 11, e1004615 (2015).
Fusco, D., Gralka, M., Kayser, J., Anderson, A. & Hallatschek, O. Excess of mutational jackpot events in expanding populations revealed by spatial Luria–Delbrück experiments. Nat. Commun. 7, 12760 (2016).
Weinstein, B. T., Lavrentovich, M. O., Möbius, W., Murray, A. W. & Nelson, D. R. Genetic drift and selection in many-allele range expansions. PLOS Comput. Biol. 13, e1005866 (2017).
Mesibov, R., Ordal, G. W. & Adler, J. The range of attractant concentrations for bacterial chemotaxis and the threshold and size of response over this range. Weber law and related phenomena. J. Gen. Physiol. 62, 203–223 (1973).
Fraebel, D. T. et al. Environment determines evolutionary trajectory in a constrained phenotypic space. eLife 6, e24669 (2017).
Blattner, F. R. et al. The complete genome sequence of Escherichia coli K-12. Science 277, 1453–1462 (1997).
Acknowledgements
We thank C. Liu and X. Fu for initiating this study, and H. Berg, P. Cluzel, K. Fahrner, J. S. Parkinson, T. Pilizota, T. Shimizu, V. Sourjik and Y. Tu for discussions. This work is supported by the NIH (R01GM95903) through T. Hwa and the NSF Program PoLS (grant 1411313) through M.V. T. Honda acknowledges a JASSO long-term graduate fellowship award.
Author information
Authors and Affiliations
Contributions
J.C., T. Honda, M.V. and T. Hwa designed this study. Experiments were performed by T. Honda and J.C., with contributions by J.W.-N. and Y.T. in characterizing swimming. J.C. and T. Hwa developed the model, and J.C. and Y.T. performed the numerical simulations. All authors contributed to the analysis of experimental and simulation data. J.C., T. Honda, Y.T., M.V. and T. Hwa participated in the writing of the paper and the Supplementary Information.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Expansion speed measurements.
a, Temporal evolution of front position for a population of E. coli HE206 cells (wild type) grown on a soft-agar plate with saturating amounts of different carbon sources (see legend) and 100 μM aspartate as attractant. Lines show linear fits. Experiments were repeated at least twice with similar results. b, Temporal evolution of front position for HE443 cells grown on 40 mM glycerol and 100 μM aspartate with different amounts of the inducer 3MBA (see legend) that titrate glycerol uptake26, resulting in different growth rates (Supplementary Table 2). The experiments were repeated at least twice with similar results. c, Expansion speed and its dependence on growth rate for the commonly used E. coli K-12 strain MG1655B77 (red symbols) and the K-12 variant RP437 (blue symbols) frequently used in motility studies (see Supplementary Text 1.1). Growth conditions were changed by varying the carbon source (from lower to higher growth rates: acetate, mannose, glycerol, glucose; see legend). Aspartate (100 μM) was added as the attractant. For the experiments with RP437, four amino acids (methionine, leucine, threonine, histidine) were provided at 1 mM each in the medium to sustain cell growth. Data from Fig. 1c are shown in grey for comparison. Data points represent the mean of two biological replicates, except for growth rates in acetate and mannose, which were from a single experiment. d, Expansion speeds plotted against the batch culture growth rate for populations grown in glycerol, glucose or aspartate as the only carbon source, without supplement of additional attractant (purple symbols). Growth on serine is very slow (<0.1 h−1) and is not shown on the plot. Expansion speeds were much slower in these media without chemoattractant supplement, even though glucose and aspartate are both attractants themselves21,45. The same was observed for a Δtar knockout strain when both glycerol and aspartate were present (open triangle). See Supplementary Tables 2, 3 for data values and sample sizes. Data from Fig. 1c are shown in grey for comparison. e, f, The difference between migration with and without additional attractant is further illustrated for growth on glycerol when growth rates are titrated (e), and for expansion when other carbon sources are provided (f). Hashed bars highlight additional increase in expansion speed (red) and growth rate (blue) when attractant is provided. In each case, supplementing low amounts of attractant(s) greatly increases expansion speed without affecting growth rate much. The graphs were created based on mean values listed in Supplementary Tables 2, 3. g, Expansion speed and its dependence on growth rate when two attractants are present (20 mM glycerol + 100 µM aspartate + 100 µM serine, green symbols) or for complex medium (CAA + carbon source, orange symbols). Data for single attractants (from Fig. 1c) are shown for comparison (20 mM glycerol + 100 µM aspartate, red; 20 mM glycerol + 100 µM serine, blue). Lines indicate fits to square-root dependencies as anticipated from a simple scaling analysis (Extended Data Fig. 9d). Data points in asp + ser (green points) represent means of two biological replicates (n = 2), except for growth rates of HE433 and HE443, which were from a single experiment (n = 1).
Extended Data Fig. 2 Swimming characteristics in liquid media (well-mixed conditions, no gradients).
a–d, Average swimming speed (purple circles) and the fraction of motile cells (grey squares) were characterized for cells taken from batch cultures along a growth curve at different optical densities (OD600; green triangles). For each condition, data points were collected from a single experiment. a, Culture was grown in LB medium, starting with an overnight LB culture that was sitting in saturation for 18 h before dilution into fresh LB medium at time zero. This experiment was essentially a repeat of previous work14,15; similar results were obtained, with motility increasing as growth progressed. Our data in the following panels suggest that most of the increase in swimming speed resulted from the increased fraction of motile cells in the first 2 h. b, Culture was grown in minimal medium with 10 mM glycerol and 1.7 mM aspartate, starting with an overnight pre-culture (same medium) that was in saturation for about 18 h before inoculation into fresh medium (time zero). As observed for LB (a), it took several hours for both the motile fraction and swimming speed to recover. c, Culture was grown in LB medium continuously for 10 generations, with bacterial density always kept below OD600 = 0.5 before dilution to fresh LB at time zero. Both the motile fraction and the swimming speeds are high in the exponential growth phase (0–2 h) except for a dip at an OD600 of approximately 0.5. Swimming speed and motile fraction decreased after the stationary phase was reached. d, Culture was grown in the same minimal medium (glycerol + aspartate) for about 20 generations, with bacterial density maintained below OD600 = 0.6 before measurement. As with LB (c), swimming speed and motile fraction remained high in the exponential growth phase (0–4 h) before sharply decreasing after entering the stationary phase. The strong variation of the fraction of motile cells observed here is in line with previous observations on cell-to-cell variation in swimming behaviour68,69 and can strongly affect the dynamics of migrating populations24,70. e, f, Swimming behaviour observed in steady-state growth (as in d for the first ~3 h) for different (relatively fast) growth conditions and different E. coli strains (Supplementary Table 5). Swimming speeds (v) and durations between tumbling events (τ) obtained from trajectory analysis are shown in e and f, respectively. Black symbols show results for the NCM3722B-derived strains (HE206, HE433, HE443; growth at 37 °C) mainly used in this study. A similar weak dependence of quantities on growth was observed for MG1655B (red symbols, growth at 37 °C) and RP437 (blue symbols, growth at 30 °C). g, Estimated effective diffusion coefficient, D = v2τ, for the different growth conditions in NCM3722B-derived strains. e–g, Data points show the means of two biological replicates for HE433 and HE443 and the results of single experiments for HE206, MG1655B and RP437. See Supplementary Text 1.2, 1.3 for methods, Supplementary Table 5 for data values and conditions, and Supplementary Text 1.1 for strain details.
Extended Data Fig. 3 Single-cell motility analysis in agar by confocal microscopy.
Thirty-second videos allowing us to track the movement of single cells were acquired (see Supplementary Video 7 for an example). a, Example of trajectories derived from cell tracking analysis. Each colour indicates the trajectory of one cell over a span of on average 75 frames (5.1 s). b, Diffusive behaviour was obtained by a linear fit of displacement variance over time (var(Δx) = 2DΔt). This analysis was performed for strain HE274 (wild type) growing in 40 mM glycerol and 100 μM aspartate (reference condition; Supplementary Text 1.5). Data shown here are for measurements in front of the expanding population (ahead of the density peak; however, the diffusion coefficient obtained at different locations does not exhibit much positional dependency, see below). Repeat of experiment showed similar results. c, Similar effective diffusion coefficients for swimming in soft-agar were obtained for other growth conditions (orange symbols; Supplementary Table 6) following the same trend as predicted from liquid culture measurements (black symbols, same as in Extended Data Fig. 2g). The diffusion measurements in soft agar were repeated twice with similar results. The data points represent means of two biological replicates. See Supplementary Table 6 for data values and conditions. d, To resolve cellular swimming behaviour of the expanding population at different spatial positions in the agar plate, videos allowing us to track single cells were acquired sequentially at a fixed position (of the agar plate) over time, for different acquisition times tacq over which videos were taken (up to several hours for each position). Image direction x was aligned with direction of migration. In this setup, the migrating population (with speed u) passes the point of acquisition at a determined time, allowing us to determine the local drift speeds and diffusion coefficients relative to the front position: x = x0 − utacq (Supplementary Text 1.5). e, Density obtained by cell counting (green line) compared to population density obtained using the approach in Extended Data Fig. 4 (fluorescence scans, red line). The spatial resolution of the latter is much coarser, each measurement point being a black dot on the red line. For comparison, the simulation result (GM model, Fig. 3) is shown in green and moderately deviates from the measured profile. f, Analysis of average displacement along x (direction of migration) and y (direction perpendicular to migration) over time for an acquisition time tacq corresponding to a position at the front bulge (x = 21.3 cm, indicated by the dashed lines in e, g, h). The average displacement (purple symbols) increased linearly in time along the direction of migration but was negligible perpendicular to the direction of migration (fitted purple lines show drift speed in each direction, ⟨Δx/Δt⟩ and ⟨Δy/Δt⟩). g, Position dependence of the drift (in the direction of expansion) was determined at different tacq, corresponding to different positions of the expanding population. For ease of reference, cellular densities (e) are shown again as green symbols. Up to the resolution of the data, the drift velocity vanished to the left of the density trough (x < 19 mm). h, Position dependence of the diffusion coefficient. Using the approach from b to determine the diffusion coefficient at different tacq, we obtained the results shown as orange symbols. A moderate (~20%) increase in D is observed at the very front of the population. This spatial dependence may be due to the accumulation of faster swimming cells at the front9. All data in e–h are from a single expansion experiment done under reference conditions (40 mM glycerol + 100 μM aspartate; 2:1 mixture of fluorescent variant HE274 and non-fluorescent variant HE339). Similar results were obtained for one biological replicate. Error bars in e, h denote s.d. and were calculated from repeated observations at three different times during the same expansion process.
Extended Data Fig. 4 Population-level observations of growth and expansion by confocal microscopy.
Densities of bacteria growing in soft agar were determined at various times using confocal microscopy and fluorescently labelled cells; see Supplementary Text 1.4. a, Calibration of fluorescence intensity. Known numbers of cells were transferred from a batch culture to a fresh but cold soft-agar plate. After agar solidification (<10 min), intensity was measured. Fitted line gives relation between observed intensity (fluorescence integrated along the agar thickness) and cell density measured in batch culture (OD600). b, Example of experiment to obtain growth rates in agar. Data are for strain HE274 (wild-type) grown in 40 mM glycerol and 200 μM aspartate. A small number of cells was mixed uniformly into fresh soft-agar plates. After agar solidification (<10 min), fluorescence intensity was observed over time. c, Derived growth curves in soft agar based on experiments as in b. Typically, there is a fast growth regime followed by a slower regime related to oxygen consumption and limitation for OD > 0.1: with oxygen running out, cells accumulate towards the agar surface and growth becomes slower. In this work, the population was always kept in the first aerobic regime. Growth rates in the first regime (coloured lines, Supplementary Table 2) were obtained by an exponential fit of the data and are comparable to those obtained in batch culture (inset). In b, c the experiment was conducted once. d, Photograph and spatiotemporal density profiles (linear intensity scale) for population expansion under reference conditions (glycerol + 100 μM aspartate; same data as in Figs. 1a, 2b). Scale bar, 2 cm. The confocal observations under reference conditions were repeated twice with similar results. e–i, Spatiotemporal density profiles (logarithmic density scale) for population expansion under different conditions, similar to those observed for the reference conditions (glycerol + 100 μM aspartate; d). Conditions are glycerol + 100 μM serine (e), glycerol + 100 μM aspartate + 100 μM serine (f), glycerol + 0.05% CAA (g), 1% tryptone broth (h), and glucose + 100 μM aspartate (i), all with strain HE274 (wild type). Colour scale bar applies to all panels. In e–i, the experiments were conducted once; expansion speeds were highly comparable to those measured manually.
Extended Data Fig. 5 Population expansion without attractant is quantitatively captured by Fisher–Kolmogorov dynamics.
The Fisher–Kolmogorov dynamics is a canonical model to describe the dynamics of expanding populations19,20. It has been successfully used to investigate the expansion and evolution of non-moving bacteria at the front of dense bacterial colonies46,71,72,73,74. Here, we probe the Fisher–Kolmogorov dynamics and its validity to describe swimming bacteria. The Fisher–Kolmogorov dynamics is driven by population growth and undirected random motion (diffusion)32,33. To compare the predictions of Fisher–Kolmogorov dynamics to the expansion of a bacterial population in the absence of a chemoattractant, we thus independently quantified growth rates and cellular diffusion for cells homogeneously distributed in soft agar (a, b). We then compared the observed migration speed and the density profile of the migrating population (for growth on glycerol as the sole carbon source, as in Fig. 2d, top) with the Fisher–Kolmogorov predictions (c, d). a, Quantification of growth by measuring the temporal density increase of a homogeneously distributed population in agar (Extended Data Fig. 4a–c, Supplementary Text 1.4). Spatially averaged density increased exponentially with growth rate λ = 0.59 h−1 for densities <0.1 OD600. For higher densities, the growth rate decreased but this regime is not important for the propagation of the front where density is low. b, Diffusion and drift of cells homogeneously distributed in soft agar. Analysis of recorded cell movement confirms the variance of position displacement to increase linearly in time (orange symbols) with diffusion constant D = 41.5 μm2 s−1 (linear fit of var(x) = 2DΔt). In comparison, the average displacement of cells (purple symbols) and the calculated drift (⟨Δx/Δt⟩, purple line) are small, indicating the absence of directed chemotactic movement. Data show average over three independent repeats (Extended Data Fig. 3, Supplementary Text 1.5). c, d, Front and spatiotemporal dynamics of an expanding population. c, Comparison of predicted expansion speed with the observed propagation of the population front. Position of the front R(t) was determined from the observed cellular densities (threshold OD600 < 0.005); it increased linearly in time, that is, R(t) = uobs t with a speed uobs = 0.62 mm h−1. Dashed line denotes predicted expansion speed calculated as \({u}_{{\rm{FK}}}=2\sqrt{\lambda \times D}=0.59\,{\rm{mm}}\,{{\rm{h}}}^{-1}\). d, Density profile of the population front. Observed density profile can be fitted to an exponential dependence \(\rho (r,t)\sim {{\rm{e}}}^{-{k}_{{\rm{o}}{\rm{b}}{\rm{s}}}(r-R(t))}\) with kobs ≈ 1.2 mm−1. Dashed line indicates the slope of the exponential density profile predicted by the Fisher–Kolmogorov equation: \({k}_{{\rm{F}}{\rm{K}}}=\sqrt{\lambda /D}=1.99\,{{\rm{m}}{\rm{m}}}^{-1}\). The discrepancy is likely to result from the low spatial resolution of the very sharp density drop; the exponential dependence of the experimental profile is defined by just three points. All experiments were conducted once with strain HE274 (wild type), using glycerol as the carbon source (no additional attractant, glycerol cannot be sensed). Growth and cell-tracking experiments were performed with uniform cell mixture in saturating glycerol conditions (40 mM). Expansion experiments were performed with 1 mM glycerol.
Extended Data Fig. 6 Different models of chemotaxis-driven migration.
To illustrate the difference among various models of chemotactic expansion, we show here simulation results of four different models. a, The classical model proposed by Keller and Segel34 creates a self-generated attractant gradient owing to attractant consumption by the migrating population. It neglects cell growth (that is, λ = 0 in equation (3) in Fig. 3a), resulting in conservation of the total number of bacteria. It also assumes that the attractant gradient could be detected with infinite precision, such that log-sensing (Weber’s law75) can be implemented by cells down to arbitrary low attractant concentrations, (that is, equation (4) with a− = 0). The latter biologically unrealistic assumption introduces a singularity that pushes all bacteria forward at a steady migration speed, which is determined by the number of cells in the population, the conserved quantity. b, The model introduced by Novick-Cohen and Segel36 fixed the singularity in the Keller–Segel model by imposing a minimal concentration for the sensing of attractant gradient (that is, equation (4) with a− > 0). Owing to the lack of cell growth, the total number of bacteria is still conserved. In this model, the density of the front bulge decays over time because once bacteria diffuse out of the front, they lose the chemotactic gradient and cannot catch up with the front. The reduction in front density reduces the migration speed, which decays steadily towards zero. c, Model including cell growth that depends on attractant concentration (nutrient = attractant). Owing to growth, population size increases over time. However, as the attractant (nutrient) is mostly consumed at the front, there is not much growth behind the front and the trailing region behind the front is mostly flat. This scenario has been realized and analysed experimentally18; see Extended Data Fig. 8 for model details and discussion. d, The GE formulated in this study (Fig. 3a), including the chemotactic effect of an attractant, together with cell growth supplied by a major nutrient source. Front propagation of cells by chemotaxis is coupled to steady growth in the trailing region (see main text and Extended Data Fig. 9). Parameter values for all models are provided in Supplementary Table 8. For simplicity, simulations shown here were solved in one dimension (non-radial). Green lines denote bacteria density, blue lines denote attractant (or sole nutrient) concentration, brown lines denote concentration of nutrients (in addition to the attractant), purple lines show local drift (equation (4)).
Extended Data Fig. 7 Aspartate uptake and further analysis of expansion dynamics.
a–c, Characterization of aspartate uptake for different growth conditions and strains. a, Aspartate uptake was determined using a colorimetric method to quantify remaining aspartate concentrations during growth (see Supplementary Text 1.2.3). In brief, change of aspartate concentration in the medium was measured during exponential growth at different cell densities (OD). Data are for growth with glycerol as the major carbon source (40 mM) and 0.8 mM initial aspartate concentration. Measurements for native aspartate uptake (wild type) and for different aspartate-uptake mutants (ΔgltJ and ΔgltP as well as the double mutant ΔgltJP). Lines show a linear fit. Uptake rate was determined by multiplying the obtained slope by the growth rate. b, The strains shown in a with different aspartate uptake rates exhibit similar growth rates. For each strain, the data points were collected from a single experiment. c, Dependence of aspartate uptake on growth rate for strains with native aspartate uptake. Growth is varied using wild-type cells (HE206, circles) grown on different sugar sources (acetate, mannose, glycerol or glucose), or by using the glpK* mutant (HE433, triangle) or the glycerol titration mutant (HE443, squares), in glycerol with different levels of the inducer 3MBA (25, 50 or 800 μM). Aspartate (0.8 mM) was provided in each case for the measurement of aspartate uptake (see a). Line shows linear fit with parameters specified in Supplementary Text 1.2.3. a, b, Data obtained for strains carrying fluorescence plasmids. c, Data obtained for non-fluorescent strains (two biological replicates, means shown). Data, strain information and medium conditions including concentrations of carbon sources are provided in Supplementary Table 7. d, Expansion dynamics with glucose as the primary carbon source. The dynamics of the front, shown to be described well by the GE model in Fig. 3b in glycerol with aspartate, is examined with the primary carbon source being glucose (20 mM). In the presence of 100 μM aspartate, the observed front propagation dynamics (purple squares) is correctly captured by the GE model again (purple line), by merely replacing the growth rate by that in glucose (λ = 1.0 h−1) with no additional adjustment of the chemotactic coefficient χ0. For reference, expansion is also shown for the condition in which no additional chemoattractant was provided (0 μM aspartate, open green squares), and the corresponding data for the condition in which glycerol was the primary carbon source (open black circles and corresponding lines from Fig. 3b). e, f, Dependence of expansion speed on attractant concentration with glycerol or glucose being the major nutrient. The increase in expansion speed at low attractant concentrations followed by decrease at higher concentrations, as previously observed43, is qualitatively captured by the GE model (dashed grey lines) in both cases. A better quantitative agreement between model and data is obtained when the linear growth term in the GE model (equation (1) in Fig. 3a) is changed to the logistic form λρ(1 − ρ/ρc). Here ρc is the carrying capacity, introduced to capture saturation of cell density in the front bulge (Supplementary Text 2.3). Predictions by the model are shown for different carrying capacities as coloured lines. In line with the strict requirement for oxygen when growing on glycerol and the observation that cells at high density accumulate at the agar surface when expanding with glycerol as major nutrient source (data not shown), the carrying capacity needed to resemble the observations is much lower for glycerol (e) than for glucose (f). Data points represent means of biological replicates (n = 2 or more) with error bars (s.d.) shown for n ≥ 3; see Supplementary Table 9 for data and sample sizes. g–i, Effect of varying aspartate uptake rate on expansion speed. The GE model predicts the expansion speed to be independent of the attractant uptake rate if all other parameters are kept fixed (g, dashed black line; Supplementary Text 2.2), with differences in attractant uptake compensated by changes in bacterial density at the front (h), such that the total rate of attractant depletion remains constant. This prediction was tested by characterizing the expansion dynamics of the aspartate-uptake mutants (strains HE506, HE552, HE555; Supplementary Text 1.1, Supplementary Tables 3, 7), which exhibited up to threefold difference in aspartate uptake (a), but only ~20% change in expansion speed (g). The small changes are readily accounted for by incorporating the small growth rate differences between these strains (b) into the GE model while keeping all other parameters fixed (g, green crosses). In addition, the aspartate-uptake mutants exhibited increasing peak densities at the front as predicted by compensation for reduced uptake (compare i, h). j–k, Predicted and observed changes in density profiles when varying the growth rate by titrating glycerol uptake in strain HE486 using 25 μM, 50 μM or 800 μM of the inducer 3MBA. For g, i, k, data were obtained from a single experiment for each strain and condition.
Extended Data Fig. 8 Expansion dynamics with the attractant being the sole nutrient source.
This is one of the scenarios of chemotaxis investigated previously18,76. Here, for comparison with the dynamics presented in the main text, we show the expansion dynamics of populations grown with glucose (a chemoattractant21) as the sole carbon source. a, For wild-type cells (HE206) spotted on 0.25% agar plate with glucose as the sole carbon source, photographs show the existence of an outer ring at the front of the expanding population for a range of glucose concentrations. Scale bars, 2 cm. The experiments were repeated once with similar results. b, Dependence of expansion speed on glucose concentration. Intuitively, reducing the glucose concentration would be expected to increase the expansion speed, as it would take less time for the population to consume the attractant. However, the circles show that reducing the glucose concentration reduced population expansion speed. Data show means of two biological replicates. c, Direct comparison of concentration dependence of expansion speeds in glucose only (open green squares), glycerol only (open black circles), glycerol or glucose with aspartate (red circles, purple squares); data for latter same as shown in Fig. 4d and Extended Data Fig. 7e, f. Expansion speed in glucose (~1–2 mm h−1) is faster than in glycerol (not an attractant) but well below the cases for which (low) amounts of attractants are supplemented. Shown data points represent means of biological replicates (n = 2 or larger), with error bars (s.d.) shown for n ≥ 3; see Supplementary Tables 9, 10 for data and sample sizes. d, e, To understand the expansion behaviour, we used confocal scans to obtain the density profiles. The ring observed in the photograph is seen as a subtle density bulge at the front bounding a flat-density interior. Note the lack of an exponential trailing region, as observed when an attractant supplement is present (Fig. 2b, Extended Data Fig. 4i, photographs in Fig. 1a). The observed density profiles are comparable with those previously found with galactose as the attractant and the major nutrient source18. Experiments here were done with wild-type cells (HE206) (a–c) and fluorescence cells (HE274) (d, e). The confocal experiments were conducted once (expansion speeds are highly comparable to those measured manually). f, To capture the observed behaviours, we modified the GE model (Fig. 3a) using only one variable a to describe the attractant/nutrient. Consumption of the growth-enabling attractant is directly coupled to the increase in density via the yield Y. g, h, Fixing model parameters using available data for growth and chemotaxis on glucose (see Supplementary Text 2.4, 2.3 with parameters used listed in Supplementary Table 4), the model generated expansion speeds (green line in b) and density profiles that capture the experimental observations well; for comparison, a coarse-grained spatial resolution similar to the experiments was used to display the profiles obtained by the simulations. i, The model output can further be understood by a scaling analysis (Supplementary Text 2.6), resulting in the simple relation u2 ∝ χ0λ (equation (E8.e)). This relation is of the same form as the result of the Fisher–Kolmogorov dynamics, \({u}_{{\rm{FK}}}=2\sqrt{D\lambda }\) (see Extended Data Fig. 5), but with the chemotactic coefficient χ0 replacing the diffusion coefficient D. j, k, The predicted dependence of u on λ and χ0 (black lines) is validated by numerical simulations of the model (blue circles). The square-root dependence of the expansion speed on the chemotactic coefficient χ0 stands in contrast to the linear dependence on χ0 when an attractant supplement is provided (Extended Data Fig. 9d) and shows that the expansion dynamics with or without the attractant supplement are two distinct classes of mathematical problem. Note that the quantitative gain in expansion speed for the case with a supplemented attractant comes not only from the change in dependence on the chemotactic coefficient from \(\sqrt{{\chi }_{0}}\) to χ0, but also from the freedom to use attractants that have large χ0 but small λ, which can be compensated by nutrients that give largerλ. Both aspartate and serine are strong attractants but poor nutrients, and are thus most potent when used in combination with a good nutrient source. Thus, separating the role of substances as nutrients and as cues not only relaxes the underlying mathematical constraint but also relaxes the biological constraint so that good attractants need not be good nutrients. These results provide an important support for the central thesis of this work, that chemotactic cells gain fitness by expanding in nutrient-replete conditions as a ‘foresighted’ navigation strategy (see main text).
Extended Data Fig. 9 Scaling analysis of expansion dynamics and illustration of the stochastic migration process.
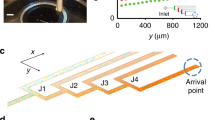
a, The exponential trailing region of the density profile is fixed by the cell growth rate λ and expansion speed u of the front. Because cells in the trailing region do not experience drift (Extended Data Fig. 3g), the apparent ‘movement’ of the trailing region at the same speed as the front bulge is possible only if it has an exponential profile, ρ(r,t) ~ ek(r − ut), with k = λ/u. b, Scaling of the expansion speed with model parameters. According to the GM model (Fig. 3a), the density peak at the propagating front is determined by a balance between cell growth and back diffusion (Fig. 4b). Using a crude scaling analysis to capture this balance, we can obtain (approximately) the quantitative determinants of the propagating speed. Consider a sharply peaked density bulge at the front, with peak density ρpeak and width w. The number of cells contained in the peak region, Npeak, is given by the relation in equation (E9.a). Cell birth rate, λNpeak, is balanced by the back-diffusion flux, which is approximated as Dρpeak/w, leading to the relation in equation (E9.b). To relate to the migration speed u, we note that around the density peak the drift speed v is nearly maximal (Fig. 4a), and equation (4) becomes \({v}_{max}\approx {\chi }_{0}\frac{{\rm{d}}}{{\rm{d}}x}\,\mathrm{ln}(a)\). In the scaling approach, we take u ~ vmax and the approximation \(\frac{{\rm{d}}}{{\rm{d}}x}\,\mathrm{ln}(a)\sim 1/w\) leading to the relation in equation (E9.c). Combining equations (E9.a) and (E9.c), we obtain equation (E9.d) with the expansion speed increasing with the square-root of the growth rate λ. Note the χ0/D factor appearing as a prefactor in the expression for u, which is responsible for the increase in the expansion speed in the presence of chemotaxis with respect to the Fisher–Kolmogorov dynamics (Extended Data Fig. 5) and for the dynamics with the attractant being the sole nutrient (Extended Data Fig. 8). c, d, Scaling results are confirmed by simulations of the GE model when varying growth rate λ (c) and chemotactic coefficient χ0 (d). e, To further illustrate the intricate dynamics at the front of the expanding population, we performed stochastic agent-based simulations looking at the trajectories of single cells. Shown here are cell trajectories for a few selected cells located within the population front (pioneers) at time t = 6.5 h. Bottom, 38 trajectories with colour indicating the time the trajectory escaped from the front and cells switched from being pioneers to being settlers, which grow and colonize localities behind the front. Red circles indicate cell division events. Highlighted area (cyan) denotes front region with aspartate concentration in the range a− < a < a+. Top, position distribution of all simulated trajectories (1,000) at time t = 8 h. See Supplementary Text 3 for details.
Extended Data Fig. 10 Modelled scenario of chemotaxis as a strict starvation response.
To examine the expansion characteristics of the population under the hypothetical scenario in which chemotaxis is a strict starvation response, we modified the GE model (Fig. 3a) to investigate the cases when chemotaxis is active either only in slow growth conditions (a, b) or within intermediate density ranges (c–e). a, To model chemotaxis being activated at slow growth, we introduced a strong dependence of chemotaxis on local growth rate (orange line). In contrast to the original GE model (dashed red line), we used a chemotactic coefficient that depends on growth conditions, χ0 = χ0(λ(n)) (orange line). Black dashed line shows growth rate in the presence of saturating glycerol. b, This dependence of chemotaxis on growth conditions leads to a marked decrease in the speed of expansion (compare orange and red dashed lines). The expansion dynamics of this model resembles the Fisher–Kolmogorov dynamics (grey dashed line), suggesting that chemotaxis does not boost population-level expansion when it is activated only under slow growth conditions. c–e, We further studied the case of swimming being a density-dependent response, active only at intermediate bacterial densities, as has been observed in batch culture measurements15 (c). Taking such a dependence of the swimming speed (v) on the local cell density (ρ) and assuming χ0 ~ v2(ρ), we looked at the expansion dynamics for several maximum values of the chemotactic coefficient (d, e). For all of the forms of χ0(ρ) shown in d, population expansion was slowed down substantially as compared to the reference case in which chemotaxis is also active at low densities (red dashed lines). The slow expansion dynamics is again similar to the Fisher–Kolmogorov dynamics, illustrating that the boost of expansion speed and population size by chemotaxis relies on chemotaxis being active at low densities. Note that in both cases analysed, we have not included the dependence of the diffusion constant on growth rate or local densities but assumed a constant value as in the original GE model. Introducing such dependences would further reduce the speed of expansion, below even that of Fisher–Kolmogorov dynamics. The origin of the slow expansion dynamics in these models is simple: a population cannot expand faster than its front, and the front is at low density and experiences the fastest growth rate.
Supplementary information
Supplementary Information
This file contains the Supplementary Tables S1 – S10 and a Supplementary Text providing an extended description of used strains, experimental methods, and the Growth Expansion Model.
41586_2019_1733_MOESM3_ESM.txt
Supplementary Data Simulation Parameters. Parameter sets for import by the custom-made Python code to numerically solve the Growth-Expansion model (Simulation code available via GitHub at https://github.com/jonascremer/chemotaxis_simulation). Provided parameter files cover all major simulation variants analyzed in the manuscript, see Supplementary Text 2.5 for details.
Video 1: Growth and expansion dynamics - glycerol as main carbon source
After initiation with a 2 μl droplet from a steadily growing cell culture (density OD600=0.2) on a soft-agar plate (x=0), the video shows the emerging density profile when 40mM glycerol is provided as carbon source. Red line shows the dynamics when 100μM aspartate was supplemented as attractant (reference condition, red line in Fig. 3A). Black line shows dynamics when no attractant was supplemented (black line in Fig. 3A). The experiment was independently repeated 3 times obtaining similar results.
Video 2: Growth and expansion dynamics – glycerol as carbon source with serine as attractant
After initiation with a 2μl droplet from a steadily growing cell culture (density OD600=0.2) on a soft-agar plate (x=0), the video shows the emerging density profile when 10mM glycerol is provided as carbon source and 100μM serine as attractant. An independent repeat obtained a similar result.
Video 3: Growth and expansion dynamics – CAA and glycerol
After initiation with a 2μl droplet from a steadily growing cell culture (density OD600=0.2) on a soft-agar plate (x=0), the video shows the emerging spatiotemporal density profile in 0.2% CAA and 40mM glycerol. The experiment was independently repeated 3 times obtaining similar results.
Video 4: Growth and expansion dynamics – TB
After initiation with a 2μl droplet from a steadily growing cell culture (density OD600=0.2) on a soft-agar plate (x=0), the video shows the emerging density profile when 1% TB is provided as nutrient source and attractant. An independent repeat showed a similar result.
Video 5: Growth and expansion dynamics – glucose as main carbon source and attractant
After initiation with a 2μl droplet from a steadily growing cell culture (density OD600=0.2) on a soft-agar plate (x=0), the video shows the emerging density profile when 5mM glucose is provided as carbon source and attractant. See also Extended Data Figure 8. An independent repeat showed a similar result.
Video 6: Modeling migration dynamics
To illustrate the migration dynamics as predicted by the model, we here show the model variables (bacterial cell density, nutrient concentration, and attractant concentration) and their variation over time. The upper panel shows these variables within the lab frame, while the lower panel shows them within a co-moving frame with the origin defined as point of maximum drift. The figure outline and colour scheme follow Fig. 4A. Cyan region denotes attractant concentration within the detectable range (a−,a+).
Video 7: Cellular swimming behaviour in soft-agar
Single cell detection and tracking using confocal microscopy allows quantification of swimming behaviour in agar; see Fig. S3 and Supplementary Text 1.5. We show here an exemplary video for cells swimming in the presence of glycerol + 100μM aspartate (reference condition). Left panel shows taken images. The observed area covers ≈ 200μm×200μm and the 100 frames shown are taken within 6.7 seconds (video plays 3 times slower than real time). Right panel shows detected cells. Emerging trajectories are shown in both panels with different colours corresponding to different cells. The experiment was independently repeated 3 times obtaining similar results and trajectory statistics.
Source data
Rights and permissions
About this article
Cite this article
Cremer, J., Honda, T., Tang, Y. et al. Chemotaxis as a navigation strategy to boost range expansion. Nature 575, 658–663 (2019). https://doi.org/10.1038/s41586-019-1733-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1733-y
This article is cited by
-
Microbes in porous environments: from active interactions to emergent feedback
Biophysical Reviews (2024)
-
Spontaneous vortex formation by microswimmers with retarded attractions
Nature Communications (2023)
-
Proliferating active matter
Nature Reviews Physics (2023)
-
Emergent behaviour and neural dynamics in artificial agents tracking odour plumes
Nature Machine Intelligence (2023)
-
Motility mediates satellite formation in confined biofilms
The ISME Journal (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.