Abstract
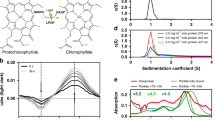
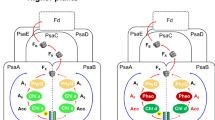
The enzyme protochlorophyllide oxidoreductase (POR) catalyses a light-dependent step in chlorophyll biosynthesis that is essential to photosynthesis and, ultimately, all life on Earth1,2,3. POR, which is one of three known light-dependent enzymes4,5, catalyses reduction of the photosensitizer and substrate protochlorophyllide to form the pigment chlorophyllide. Despite its biological importance, the structural basis for POR photocatalysis has remained unknown. Here we report crystal structures of cyanobacterial PORs from Thermosynechococcus elongatus and Synechocystis sp. in their free forms, and in complex with the nicotinamide coenzyme. Our structural models and simulations of the ternary protochlorophyllide–NADPH–POR complex identify multiple interactions in the POR active site that are important for protochlorophyllide binding, photosensitization and photochemical conversion to chlorophyllide. We demonstrate the importance of active-site architecture and protochlorophyllide structure in driving POR photochemistry in experiments using POR variants and protochlorophyllide analogues. These studies reveal how the POR active site facilitates light-driven reduction of protochlorophyllide by localized hydride transfer from NADPH and long-range proton transfer along structurally defined proton-transfer pathways.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Yang, J. & Cheng, Q. Origin and evolution of the light-dependent protochlorophyllide oxidoreductase (LPOR) genes. Plant Biol. 6, 537–544 (2004).
Scrutton, N. S., Groot, M. L. & Heyes, D. J. Excited state dynamics and catalytic mechanism of the light-driven enzyme protochlorophyllide oxidoreductase. Phys. Chem. Chem. Phys. 14, 8818–8824 (2012).
Gabruk, M. & Mysliwa-Kurdziel, B. Light-dependent protochlorophyllide oxidoreductase: phylogeny, regulation, and catalytic properties. Biochemistry 54, 5255–5262 (2015).
Aubert, C., Vos, M. H., Mathis, P., Eker, A. P. & Brettel, K. Intraprotein radical transfer during photoactivation of DNA photolyase. Nature 405, 586–590 (2000).
Sorigué, D. et al. An algal photoenzyme converts fatty acids to hydrocarbons. Science 357, 903–907 (2017).
Dietzek, B. et al. Excited-state processes in protochlorophyllide a a femtosecond time-resolved absorption study. Chem. Phys. Lett. 397, 110–115 (2004).
Dietzek, B. et al. Dynamics of charge separation in the excited-state chemistry of protochlorophyllide. Chem. Phys. Lett. 492, 157–163 (2010).
Sytina, O. A. et al. Protochlorophyllide excited-state dynamics in organic solvents studied by time-resolved visible and mid-infrared spectroscopy. J. Phys. Chem. B 114, 4335–4344 (2010).
Zhao, G. J. & Han, K. L. Site-specific solvation of the photoexcited protochlorophyllide a in methanol: formation of the hydrogen-bonded intermediate state induced by hydrogen-bond strengthening. Biophys. J. 94, 38–46 (2008).
Heyes, D. J. et al. Excited-state charge separation in the photochemical mechanism of the light-driven enzyme protochlorophyllide oxidoreductase. Angew. Chem. Int. Ed. 54, 1512–1515 (2015).
Heyes, D. J. et al. Excited-state properties of protochlorophyllide analogues and implications for light-driven synthesis of chlorophyll. J. Phys. Chem. B 121, 1312–1320 (2017).
Brandariz-de-Pedro, G. et al. Direct evidence of an excited-state triplet species upon photoactivation of the chlorophyll precursor protochlorophyllide. J. Phys. Chem. Lett. 8, 1219–1223 (2017).
Heyes, D. J. et al. The first catalytic step of the light-driven enzyme protochlorophyllide oxidoreductase proceeds via a charge transfer complex. J. Biol. Chem. 281, 26847–26853 (2006).
Heyes, D. J., Sakuma, M., de Visser, S. P. & Scrutton, N. S. Nuclear quantum tunneling in the light-activated enzyme protochlorophyllide oxidoreductase. J. Biol. Chem. 284, 3762–3767 (2009).
Heyes, D. J., Sakuma, M. & Scrutton, N. S. Solvent-slaved protein motions accompany proton but not hydride tunneling in light-activated protochlorophyllide oxidoreductase. Angew. Chem. Int. Ed. 48, 3850–3853 (2009).
Heyes, D. J., Levy, C., Sakuma, M., Robertson, D. L. & Scrutton, N. S. A twin-track approach has optimized proton and hydride transfer by dynamically coupled tunneling during the evolution of protochlorophyllide oxidoreductase. J. Biol. Chem. 286, 11849–11854 (2011).
Hoeven, R., Hardman, S. J. O., Heyes, D. J. & Scrutton, N. S. Cross-species analysis of protein dynamics associated with hydride and proton transfer in the catalytic cycle of the light-driven enzyme protochlorophyllide oxidoreductase. Biochemistry 55, 903–913 (2016).
Archipowa, N., Kutta, R. J., Heyes, D. J. & Scrutton, N. S. Stepwise hydride transfer in a biological system: insights into the reaction mechanism of the light-dependent protochlorophyllide oxidoreductase. Angew. Chem. Int. Ed. 57, 2682–2686 (2018).
Kavanagh, K. L., Jörnvall, H., Persson, B. & Oppermann, U. Medium- and short-chain dehydrogenase/reductase gene and protein families : the SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes. Cell. Mol. Life Sci. 65, 3895–3906 (2008).
Korman, T. P., Tan, Y. H., Wong, J., Luo, R. & Tsai, S. C. Inhibition kinetics and emodin cocrystal structure of a type II polyketide ketoreductase. Biochemistry 47, 1837–1847 (2008).
Javidpour, P. et al. The determinants of activity and specificity in actinorhodin type II polyketide ketoreductase. Chem. Biol. 20, 1225–1234 (2013).
Blaise, M., Van Wyk, N., Banères-Roquet, F., Guérardel, Y. & Kremer, L. Binding of NADP+ triggers an open-to-closed transition in a mycobacterial FabG β-ketoacyl-ACP reductase. Biochem. J. 474, 907–921 (2017).
Zhao, F. J. et al. Crystal structure and iterative saturation mutagenesis of ChKRED20 for expanded catalytic scope. Appl. Microbiol. Biotechnol. 101, 8395–8404 (2017).
Menon, B. R. K., Hardman, S. J. O., Scrutton, N. S. & Heyes, D. J. Multiple active site residues are important for photochemical efficiency in the light-activated enzyme protochlorophyllide oxidoreductase (POR). J. Photochem. Photobiol. B 161, 236–243 (2016).
Klement, H., Helfrich, M., Oster, U., Schoch, S. & Rüdiger, W. Pigment-free NADPH: protochlorophyllide oxidoreductase from Avena sativa L. Eur. J. Biochem. 265, 862–874 (1999).
Menon, B. R. K., Waltho, J. P., Scrutton, N. S. & Heyes, D. J. Cryogenic and laser photoexcitation studies identify multiple roles for active site residues in the light-driven enzyme protochlorophyllide oxidoreductase. J. Biol. Chem. 284, 18160–18166 (2009).
Ashkenazy, H. et al. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344–W350 (2016).
Acknowledgements
The work was funded by the Engineering and Physical Sciences Research Council (fellowship to N.S.S., EP/J020192/1). We thank Diamond Light Source beamlines I03 & I04 (proposal numbers MX8997-35, MX12788-8, 19, 42 and 62) and Shanghai Synchrotron Radiation Facility beamlines BL17U1 and BL19U1 for assistance during data collection. Time-resolved visible absorption measurements were performed using instrumentation funded by BBSRC Alert14 Award BB/M011658/1. This work was also supported by MOST 973 Project Biological Nitrogen Fixation (2010CB126504), National Basic Research Program of China (2014CB910304) and National Natural Science Foundation of China (31230004 and 81572090). We thank R. Dixon and R. Read for early discussions and support, and C. E. Bauer and Y. Fujita for kindly providing the strain ZY5 for the production of Pchlide.
Author information
Authors and Affiliations
Contributions
D.J.H., A.Z., Q.C. and N.S.S. initiated and coordinated the project. S.Z., Q.C., A.Z., N.S.S., D.L. and D.J.H. designed experiments, analysed data and wrote the manuscript with contributions from other authors. S.Z., M.S., L.F., W.S., H.L., J.Y. and X.L. produced and crystallized the proteins. A.Z. and C.W.L. collected and processed diffraction data and solved the structures. S.H. and L.O.J. performed the docking and molecular dynamics simulations. S.J.O.H. and D.J.H. performed time-resolved spectroscopy measurements. R.H. and M.S. assisted with protein purification and characterization of POR variants. X.Y., M.L. and Z.R. advised on all aspects. All authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Peer review information Nature thanks Marco De Vivo, Sibongile Mafu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
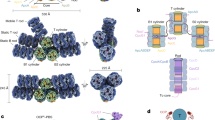
Extended Data Fig. 1 Crystal structures of the T. elongatus and Synechocystis PORs.
a, T. elongatus apo-POR. b, T. elongatus POR with bound NADPH. c, Synechocystis POR with bound NADPH. The protein structure is coloured according to secondary structure; cyan, α-helix; magenta, β-sheet; green, loop. d, Alignment of the POR crystal structures that we solve here. Magenta, apo structure of T. elongatus POR; yellow, structure of NADPH-bound T. elongatus POR; blue, structure of NADPH-bound Synechocystis POR; orange, missing loops in T. elongatus POR (which are present in the crystal structure of the Synechocystis POR). e, Evolutionary conservation of Synechocystis POR structure. Evolutionary conservation of amino acid positions in the Synechocystis POR protein has been performed on the basis of the phylogenetic relations between homologous sequences, using the online ConSurf server27.
Extended Data Fig. 2 Comparison of the structure of the Synechocystis POR structure with that of the SDR-family protein β-ketoacyl reductase (PDB code 2B4Q).
a, Overall structure of β-ketoacyl reductase protein (coloured by secondary structure). b, Alignment of the Synechocystis POR (blue) and β-ketoacyl reductase (orange). NADPH is shown as a red stick.
Extended Data Fig. 3 Molecular-dynamics simulations of the T. elongatus POR.
a–c, Overlays of structures from unrestrained molecular-dynamics simulations at 500-ps intervals for apo enzyme (a), POR–NADPH complex (b) and the ternary complex (c). d, Root-mean-squared deviation (RMSD) versus time (top) and the per-residue root-mean squared fluctuation (RMSF) (bottom), calculated for the non-hydrogen atoms of the protein. e, Distance distributions for the Pchlide-binding coordinate R0, and the distances between the hydride donor and acceptor atoms (R1) and the proton donor and acceptor atoms (R2).
Extended Data Fig. 4 Electron density map for NADPH bound to the T. elongatus and Synechocystis PORs.
a, NADPH anaerobic soaking with the T. elongatus POR crystal. A low-resolution 3.5 Å electron density map (field-emission microscopy) contoured at 1σ (green mesh) along with an Fo − Fc omit map contoured at 4σ (magenta mesh) is shown for the NADPH region of an anaerobically soaked T. elongatus POR crystal. An all-atom coloured stick representation of the aerobically soaked high-resolution NADPH–POR structure and associated active-site interactions are shown along with a stick representation of the anaerobically soaked NADPH in green. b, Structure of the NADPH-binding site of the Synechocystis POR. Hydrogen bonds between key residues and NADPH are shown as red dashes. The electron density for NADPH (omit Fo − Fc map contoured at 3σ) is coloured green.
Extended Data Fig. 5 Modelling of the T. elongatus POR–Pchlide–NADPH ternary complex.
a–c, Three stages of modelling the T. elongatus POR–Pchlide–NADPH ternary complex. The structures are aligned and superimposed. Each panel highlights one of the three structures in blue (with the other two shown in grey): crystal structure with the chosen docked Pchlide structure (a); the structure after 20-ns annealing (b); and the final representative structure (c). The flexible residues during docking are shown as a yellow ball-and-stick representation, and the Pchlide molecule is shown in green stick representation. d, Potential of mean force (PMF) calculated by umbrella sampling as a function of R0 (Supplementary Methods) for 50-ns umbrella sampling (black), as well as for the first 20 ns (blue) and the last 20 ns (red). e, Population distributions for each bin, each sampled for 50 ns. f, Potential of mean force was calculated by umbrella sampling as a function of R0 (distance between Pchlide Mg2+ and lower edge of the POR binding pocket) (Supplementary Methods). The dip in the potential of mean force at an R0 of about 17 Å corresponds to the formation of a hydrogen bond between Y223 and the Pchlide keto group.
Extended Data Fig. 6 Configuration of the active site and donor–acceptor distance of the T. elongatus POR–NADPH–Pchlide ternary complex model.
a, Active site of the T. elongatus POR–NADPH–Pchlide ternary complex. The hydrogen-bonding network around the Pchlide and NADPH molecules is shown as red dashes. The donor–acceptor distance for hydride and proton transfer is shown as blue dashes. Water molecules are shown as red balls. b, View of the active site of the T. elongatus POR, highlighting the donor–acceptor distance for hydride transfer (shown as a blue dashed line). c, View of the active site of the T. elongatus POR, highlighting the donor–acceptor distance for proton transfer (shown as a blue dashed line). d, Summary of the activity, binding and inhibition data for the Pchlide analogues. The structures and apparent kcat, Km, Kd and Ki (where applicable, in each case) are shown for Pchlide (I), protopheophorbide (II), Pchlide with a C-17 methylester (III), Pchlide with a C-13–OH (IV) and Pchlide with a C-13 and C-15 methylester (V). The red circles show the regions of the Pchlide molecule that have previously been shown to be important for activity (central Mg2+, ring E and the side chain at the C-17 position). The structures of all of the Pchlide derivatives described in the present study are shown with the modifications indicated by dashed red circles.
Extended Data Fig. 7 Hydrogen-bonding interactions between Tyr223 and the C-13 keto group during Pchlide binding.
a–g, Change in hydrogen bonding between Tyr223 and Pchlide C-13 keto group during Pchlide binding. Plots of the distance R0 between Pchlide Mg2+ and base of POR binding pocket (black), and the distance RHB (HB, hydrogen bonding) between the Tyr223 hydroxy proton and keto oxygen (blue) from 20-ns molecular dynamics simulations at increasing R0 values. h, Average RHB for each 0.5 Å bin for R0. i, Extended 100-ns molecular-dynamics simualation with R0 restrained at 1.7 Å to further illustrate the stability of the hydrogen bond between Y223 and the keto group. j–m, As the Pchlide leaves the binding pocket (shown sequentially from j to m), the Tyr 223 residue is free to move around and form a transient hydrogen bond with the C-13 keto group of Pchlide to ‘guide’ the Pchlide into its final orientation. The protein backbone is shown as a grey cartoon. At each stage, the Pchlide and Y223 molecules have been highlighted with thicker sticks in the figure.
Extended Data Fig. 8 Evolution associated difference spectra that result from global analysis using a sequential model of visible transient absorption data collected between 0.6 ns and 2.7 μs.
a–h, Evolution associated difference spectra (EADS) are shown for Pchlide (a), POR with NADPH and Pchlide (b), protopheophorbide (c), POR with NADPH and protopheophorbide (d), Pchlide with a C-13 and C-15 methylester (e), POR with NADPH, and Pchlide with a C-13 and C-15 methylester (f), Pchlide with a C-13–OH (g), POR with NADPH, and Pchlide with a C-13–OH (h). All data could be fitted using two EADS, except for POR with NADPH and Pchlide, which required three EADS owing to the formation of the hydride transfer intermediate (spectrum in red in b). The absence of any additional intermediates for the Pchlide analogues in the presence of POR implies impaired photochemistry.
Extended Data Fig. 9 Potential interactions in the T. elongatus POR ternary complex model.
a, The Pchlide molecule was chosen as the ligand, and the POR protein and solvent were chosen as the receptor. The 2D interaction map was calculated through molecular operating environment software (Chemical Computing Group). b, The NADPH molecule was chosen as the ligand, and the POR protein and solvent were chosen as the receptor. The 2D interaction map was calculated through molecular operating environment software (Chemical Computing Group).
Supplementary information
Supplementary Information
This file contains Supplementary Methods, Supplementary Figures 1–13 and Supplementary Table.
Video 1
Protochlorophyllide binding video.
Rights and permissions
About this article
Cite this article
Zhang, S., Heyes, D.J., Feng, L. et al. Structural basis for enzymatic photocatalysis in chlorophyll biosynthesis. Nature 574, 722–725 (2019). https://doi.org/10.1038/s41586-019-1685-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1685-2
This article is cited by
-
Genetic dissection of maize (Zea mays L.) chlorophyll content using multi-locus genome-wide association studies
BMC Genomics (2023)
-
CATase-immobilized hydrogel platform molded by photo-enzyme coupling polymerization for effectively preventing postoperative abdominal adhesion
Science China Chemistry (2023)
-
Dunaliella Ds-26-16 acts as a global regulator to enhance salt tolerance by coordinating multiple responses in Arabidopsis seedlings
Planta (2023)
-
Photocatalytic dehydrogenative C-C coupling of acetonitrile to succinonitrile
Nature Communications (2022)
-
A photonanozyme with light-empowered specific peroxidase-mimicking activity
Nano Research (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.