Abstract
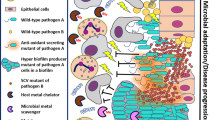
Many microbial communities, including those involved in chronic human infections, are patterned at the micron scale. In this Review, we summarize recent work that has defined the spatial arrangement of microorganisms in infection and begun to demonstrate how changes in spatial patterning correlate with disease. Advances in microscopy have refined our understanding of microbial micron-scale biogeography in samples from humans. These findings then serve as a benchmark for studying the role of spatial patterning in preclinical models, which provide experimental versatility to investigate the interplay between biogeography and pathogenesis. Experimentation using preclinical models has begun to show how spatial patterning influences the interactions between cells, their ability to coexist, their virulence and their recalcitrance to treatment. Future work to study the role of biogeography in infection and the functional biogeography of microorganisms will further refine our understanding of the interplay of spatial patterning, pathogen virulence and disease outcomes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Hall-Stoodley, L., Costerton, J. W. & Stoodley, P. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2, 95–108 (2004).
Flemming, H. C. & Wuertz, S. Bacteria and archaea on Earth and their abundance in biofilms. Nat. Rev. Microbiol. 17, 247–260 (2019).
Azimi, S., Klementiev, A. D., Whiteley, M. & Diggle, S. P. Bacterial quorum sensing during infection. Annu. Rev. Microbiol. 74, 201–219 (2020).
Ibberson, C. B. & Whiteley, M. The social life of microbes in chronic infection. Curr. Opin. Microbiol. 53, 44–50 (2020).
Martiny, J. B. et al. Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 4, 102–112 (2006).
Nemergut, D. R. et al. Global patterns in the biogeography of bacterial taxa. Env. Microbiol. 13, 135–144 (2011).
Whiteley, M., Lee, K. M. & Greenberg, E. P. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA 96, 13904–13909 (1999).
Giraudo, A. T., Mansilla, C., Chan, A., Raspanti, C. & Nagel, R. Studies on the expression of regulatory locus sae in Staphylococcus aureus. Curr. Microbiol. 46, 246–250 (2003).
Ibberson, C. B. et al. Co-infecting microorganisms dramatically alter pathogen gene essentiality during polymicrobial infection. Nat. Microbiol. 2, 17079 (2017).
Cornforth, D. M. et al. Pseudomonas aeruginosa transcriptome during human infection. Proc. Natl Acad. Sci. USA 115, E5125–E5134 (2018).
Stacy, A. et al. Bacterial fight-and-flight responses enhance virulence in a polymicrobial infection. Proc. Natl Acad. Sci. USA 111, 7819–7824 (2014). In this pivotal study, the authors show that precise spatial patterning impacts virulence and characterize the metabolic and genetic factors involved in this relationship.
Kolenbrander, P. E., Egland, P. G., Diaz, P. I. & Palmer, R. J. Jr. Genome–genome interactions: bacterial communities in initial dental plaque. Trends Microbiol. 13, 11–15 (2005).
Whiteley, M., Diggle, S. P. & Greenberg, E. P. Progress in and promise of bacterial quorum sensing research. Nature 551, 313–320 (2017).
Bassler, B. L. How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr. Opin. Microbiol. 2, 582–587 (1999).
Williams, P. et al. Quorum sensing and the population-dependent control of virulence. Phil. Trans. R. Soc. Lond. B 355, 667–680 (2000).
Tegtmeyer, N., Wessler, S. & Backert, S. Role of the cag-pathogenicity island encoded type IV secretion system in Helicobacter pylori pathogenesis. FEBS J. 278, 1190–1202 (2011).
Zhou, Y. et al. Hcp family proteins secreted via the type VI secretion system coordinately regulate Escherichia coli K1 interaction with human brain microvascular endothelial cells. Infect. Immun. 80, 1243–1251 (2012).
Aubert, D. F. et al. A Burkholderia type VI effector deamidates Rho GTPases to activate the pyrin inflammasome and trigger inflammation. Cell Host Microbe 19, 664–674 (2016).
Shalom, G., Shaw, J. G. & Thomas, M. S. In vivo expression technology identifies a type VI secretion system locus in Burkholderia pseudomallei that is induced upon invasion of macrophages. Microbiology 153, 2689–2699 (2007).
Stacy, A., McNally, L., Darch, S. E., Brown, S. P. & Whiteley, M. The biogeography of polymicrobial infection. Nat. Rev. Microbiol. 14, 93–105 (2016). This review provides a primer on the factors that drive microbiogeography during infection, including attachment, the physiochemical environment, host factors and polymicrobial interactions.
Alhede, M. et al. Combination of microscopic techniques reveals a comprehensive visual impression of biofilm structure and composition. FEMS Immunol. Med. Microbiol. 65, 335–342 (2012).
Hughes, C. V., Kolenbrander, P. E., Andersen, R. N. & Moore, L. V. Coaggregation properties of human oral Veillonella spp.: relationship to colonization site and oral ecology. Appl. Env. Microbiol. 54, 1957–1963 (1988).
Werner, E. et al. Stratified growth in Pseudomonas aeruginosa biofilms. Appl. Env. Microbiol. 70, 6188–6196 (2004).
Rogers, J. D., Palmer, R. J. Jr., Kolenbrander, P. E. & Scannapieco, F. A. Role of Streptococcus gordonii amylase-binding protein A in adhesion to hydroxyapatite, starch metabolism, and biofilm formation. Infect. Immun. 69, 7046–7056 (2001).
Angelichio, M. J., Spector, J., Waldor, M. K. & Camilli, A. Vibrio cholerae intestinal population dynamics in the suckling mouse model of infection. Infect. Immun. 67, 3733–3739 (1999).
Earle, K. A. et al. Quantitative imaging of gut microbiota spatial organization. Cell Host Microbe 18, 478–488 (2015).
McAlester, G., O’Gara, F. & Morrissey, J. P. Signal-mediated interactions between Pseudomonas aeruginosa and Candida albicans. J. Med. Microbiol. 57, 563–569 (2008).
Reddinger, R. M., Luke-Marshall, N. R., Sauberan, S. L., Hakansson, A. P. & Campagnari, A. A. Streptococcus pneumoniae modulates Staphylococcus aureus biofilm dispersion and the transition from colonization to invasive disease. mBio 9, e02089-17 (2018).
Schulte, M. & Hensel, M. Models of intestinal infection by Salmonella enterica: introduction of a new neonate mouse model. F1000Res. 5, 1498 (2016).
Frank, R. M. & Houver, G. in Dental Plaque (ed. McHugh, W. D.) 85–108 (S Livingstone, 1970).
Kharazmi, A., Giwercman, B. & Hoiby, N. Robbins device in biofilm research. Methods Enzymol. 310, 207–215 (1999).
Marrie, T. J. & Costerton, J. W. Mode of growth of bacterial pathogens in chronic polymicrobial human osteomyelitis. J. Clin. Microbiol. 22, 924–933 (1985).
Gristina, A. G. & Costerton, J. W. Bacterial adherence to biomaterials and tissue. The significance of its role in clinical sepsis. J. Bone Jt. Surg. Am. 67, 264–273 (1985).
DeLong, E. F., Taylor, L. T., Marsh, T. L. & Preston, C. M. Visualization and enumeration of marine planktonic archaea and bacteria by using polyribonucleotide probes and fluorescent in situ hybridization. Appl. Env. Microbiol. 65, 5554–5563 (1999).
DeLong, E. F., Wickham, G. S. & Pace, N. R. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science 243, 1360–1363 (1989).
Pernthaler, A., Pernthaler, J. & Amann, R. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Env. Microbiol. 68, 3094–3101 (2002).
Stoecker, K., Dorninger, C., Daims, H. & Wagner, M. Double labeling of oligonucleotide probes for fluorescence in situ hybridization (DOPE-FISH) improves signal intensity and increases rRNA accessibility. Appl. Env. Microbiol. 76, 922–926 (2010).
Valm, A. M. et al. Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proc. Natl Acad. Sci. USA 108, 4152–4157 (2011).
Valm, A. M., Oldenbourg, R. & Borisy, G. G. Multiplexed spectral imaging of 120 different fluorescent labels. PLoS ONE 11, e0158495 (2016).
Cohen-Cymberknoh, M., Kerem, E., Ferkol, T. & Elizur, A. Airway inflammation in cystic fibrosis: molecular mechanisms and clinical implications. Thorax 68, 1157–1162 (2013).
Boucher, R. C. Airway surface dehydration in cystic fibrosis: pathogenesis and therapy. Annu. Rev. Med. 58, 157–170 (2007).
Bjarnsholt, T. et al. Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr. Pulmonol. 44, 547–558 (2009). This study combines numerous techniques to visualize P. aeruginosa in the cystic fibrosis lung and in sputum, relative to immune cells.
Alhede, M. et al. The origin of extracellular DNA in bacterial biofilm infections in vivo. Pathog. Dis. 78, ftaa018 (2020).
Treweek, J. B. et al. Whole-body tissue stabilization and selective extractions via tissue–hydrogel hybrids for high-resolution intact circuit mapping and phenotyping. Nat. Protoc. 10, 1860–1896 (2015).
Yang, B. et al. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell 158, 945–958 (2014).
DePas, W. H. et al. Exposing the three-dimensional biogeography and metabolic states of pathogens in cystic fibrosis sputum via hydrogel embedding, clearing, and rRNA labeling. mBio 7, e00796-1 (2016). This study demonstrates that the use of tissue clearing and hybridization chain reaction probes allows for detecting physiological heterogeneity and spatial organization of multiple species in cystic fibrosis sputum.
Rogers, G. B., Taylor, S. L., Hoffman, L. R. & Burr, L. D. The impact of CFTR modulator therapies on CF airway microbiology. J. Cyst. Fibros. 19, 359–364 (2020).
Zhao, G. et al. Delayed wound healing in diabetic (db/db) mice with Pseudomonas aeruginosa biofilm challenge: a model for the study of chronic wounds. Wound Repair Regen. 18, 467–477 (2010).
Bjarnsholt, T. et al. Why chronic wounds will not heal: a novel hypothesis. Wound Repair Regen. 16, 2–10 (2008).
Thuenauer, R. et al. The Pseudomonas aeruginosa lectin LecB causes integrin internalization and inhibits epithelial wound healing. mBio 11, e03260-19 (2020).
Brothers, K. M. et al. Putting on the brakes: bacterial impediment of wound healing. Sci. Rep. 5, 14003 (2015).
Sen, C. K. Human wounds and its burden: an updated compendium of estimates. Adv. Wound Care 8, 39–48 (2019).
Kirketerp-Moller, K. et al. Distribution, organization, and ecology of bacteria in chronic wounds. J. Clin. Microbiol. 46, 2717–2722 (2008).
Fazli, M. et al. Nonrandom distribution of Pseudomonas aeruginosa and Staphylococcus aureus in chronic wounds. J. Clin. Microbiol. 47, 4084–4089 (2009).
Bay, L. et al. Bacterial aggregates establish at the edges of acute epidermal wounds. Adv. Wound Care 7, 105–113 (2018).
MacLeod, A. S. & Mansbridge, J. N. The innate immune system in acute and chronic wounds. Adv. Wound Care 5, 65–78 (2016).
Mark Welch, J. L., Ramirez-Puebla, S. T. & Borisy, G. G. Oral microbiome geography: micron-scale habitat and niche. Cell Host Microbe 28, 160–168 (2020).
Palmer, R. J. Jr. et al. Retrieval of biofilms from the oral cavity. Methods Enzymol. 337, 393–403 (2001).
Jones, S. J. A special relationship between spherical and filamentous microorganisms in mature human dental plaque. Arch. Oral. Biol. 17, 613–616 (1972).
Mark Welch, J. L., Rossetti, B. J., Rieken, C. W., Dewhirst, F. E. & Borisy, G. G. Biogeography of a human oral microbiome at the micron scale. Proc. Natl Acad. Sci. USA 113, E791–E800 (2016). In this article, the authors use CLASI-FISH imaging to visualize the complex microbial consortium in human dental plaque and propose a model of the plaque microbial community microbiogeography.
Wilbert, S. A., Mark Welch, J. L. & Borisy, G. G. Spatial ecology of the human tongue dorsum microbiome. Cell Rep. 30, 4003–4015 (2020).
Mark Welch, J. L., Dewhirst, F. E. & Borisy, G. G. Biogeography of the oral microbiome: the site-specialist hypothesis. Annu. Rev. Microbiol. 73, 335–358 (2019).
Carpenter, G. H. Salivary factors that maintain the normal oral commensal microflora. J. Dent. Res. 99, 644–649 (2020).
Kolenbrander, P. E. et al. Bacterial interactions and successions during plaque development. Periodontol 2000 42, 47–79 (2006).
Kim, D. et al. Spatial mapping of polymicrobial communities reveals a precise biogeography associated with human dental caries. Proc. Natl Acad. Sci. USA 117, 12375-12386 (2020). This study combines imaging of specimens from the human oral cavity, preclinical models and quantification of microbiogeography to link spatial patterning with disease.
Dufrene, Y. F. et al. Imaging modes of atomic force microscopy for application in molecular and cell biology. Nat. Nanotechnol. 12, 295–307 (2017).
Connell, J. L., Kim, J., Shear, J. B., Bard, A. J. & Whiteley, M. Real-time monitoring of quorum sensing in 3D-printed bacterial aggregates using scanning electrochemical microscopy. Proc. Natl Acad. Sci. USA 111, 18255–18260 (2014).
Klementiev, A. D., Jin, Z. & Whiteley, M. Micron scale spatial measurement of the O2 gradient surrounding a bacterial biofilm in real time. mBio 11, e02536-20 (2020).
Garg, N. et al. Three-dimensional microbiome and metabolome cartography of a diseased human lung. Cell Host Microbe 22, 705–716.e4 (2017).
Wagner, M. & Horn, H. Optical coherence tomography in biofilm research: a comprehensive review. Biotechnol. Bioeng. 114, 1386–1402 (2017).
Sussulini, A., Becker, J. S. & Becker, J. S. Laser ablation ICP-MS: application in biomedical research. Mass. Spectrom. Rev. 36, 47–57 (2017).
Zhang, P., Chen, Y. P., Qiu, J. H., Dai, Y. Z. & Feng, B. Imaging the microprocesses in biofilm matrices. Trends Biotechnol. 37, 214–226 (2019).
Caniglia, G. & Kranz, C. Scanning electrochemical microscopy and its potential for studying biofilms and antimicrobial coatings. Anal. Bioanal. Chem. 412, 6133–6148 (2020).
Skaar, E. P. Imaging infection across scales of size: from whole animals to single molecules. Annu. Rev. Microbiol. 75, 407–426 (2021). This review provides an overview of imaging methods for various infection models.
Kara, D., Luppens, S. B., van Marle, J., Özok, R. & ten Cate, J. M. Microstructural differences between single-species and dual-species biofilms of Streptococcus mutans and Veillonella parvula, before and after exposure to chlorhexidine. FEMS Microbiol. Lett. 271, 90–97 (2007).
Palmer, R. J. Jr., Diaz, P. I. & Kolenbrander, P. E. Rapid succession within the Veillonella population of a developing human oral biofilm in situ. J. Bacteriol. 188, 4117–4124 (2006).
Roberts, A. E., Kragh, K. N., Bjarnsholt, T. & Diggle, S. P. The limitations of in vitro experimentation in understanding biofilms and chronic infection. J. Mol. Biol. 427, 3646–3661 (2015).
Cornforth, D. M., Diggle, F. L., Melvin, J. A., Bomberger, J. M. & Whiteley, M. Quantitative framework for model evaluation in microbiology research using Pseudomonas aeruginosa and cystic fibrosis infection as a test case. mBio 11, e03042-19 (2020). In this study, the authors provide a framework for quantifying the accuracy of preclinical models, relative to human infections.
Ibberson, C. B. & Whiteley, M. The Staphylococcus aureus transcriptome during cystic fibrosis lung infection. mBio 10, e02774-19 (2019).
Kwiecinski, J. M. & Horswill, A. R. Staphylococcus aureus bloodstream infections: pathogenesis and regulatory mechanisms. Curr. Opin. Microbiol. 53, 51–60 (2020).
Paulsson, M., Su, Y. C., Ringwood, T., Udden, F. & Riesbeck, K. Pseudomonas aeruginosa uses multiple receptors for adherence to laminin during infection of the respiratory tract and skin wounds. Sci. Rep. 9, 18168 (2019).
Kavanaugh, J. S. et al. Identification of extracellular DNA-binding proteins in the biofilm matrix. mBio 10, e01137-19 (2019).
Moscoso, M., Garcia, E. & Lopez, R. Biofilm formation by Streptococcus pneumoniae: role of choline, extracellular DNA, and capsular polysaccharide in microbial accretion. J. Bacteriol. 188, 7785–7795 (2006).
Moser, C. et al. Novel experimental Pseudomonas aeruginosa lung infection model mimicking long-term host–pathogen interactions in cystic fibrosis. APMIS 117, 95–107 (2009).
Darch, S. E. et al. Spatial determinants of quorum signaling in a Pseudomonas aeruginosa infection model. Proc. Natl Acad. Sci. USA 115, 4779–4784 (2018).
Palmer, K. L., Aye, L. M. & Whiteley, M. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J. Bacteriol. 189, 8079–8087 (2007).
Turner, K. H., Wessel, A. K., Palmer, G. C., Murray, J. L. & Whiteley, M. Essential genome of Pseudomonas aeruginosa in cystic fibrosis sputum. Proc. Natl Acad. Sci. USA 112, 4110–4115 (2015).
Darch, S. E. et al. Phage inhibit pathogen dissemination by targeting bacterial migrants in a chronic infection model. mBio 8, e00240-17 (2017).
Barraza, J. P. & Whiteley, M. A Pseudomonas aeruginosa antimicrobial affects the biogeography but not fitness of Staphylococcus aureus during coculture. mBio 12, e00047-21 (2021).
Palmer, K. L., Mashburn, L. M., Singh, P. K. & Whiteley, M. Cystic fibrosis sputum supports growth and cues key aspects of Pseudomonas aeruginosa physiology. J. Bacteriol. 187, 5267–5277 (2005).
Azimi, S. et al. O-Specific antigen-dependent surface hydrophobicity mediates aggregate assembly type in Pseudomonas aeruginosa. mBio 12, e00860-21 (2021).
Sun, Y., Dowd, S. E., Smith, E., Rhoads, D. D. & Wolcott, R. D. In vitro multispecies Lubbock chronic wound biofilm model. Wound Repair Regen. 16, 805–813 (2008).
DeLeon, S. et al. Synergistic interactions of Pseudomonas aeruginosa and Staphylococcus aureus in an in vitro wound model. Infect. Immun. 82, 4718–4728 (2014).
Machan, Z. A., Taylor, G. W., Pitt, T. L., Cole, P. J. & Wilson, R. 2-Heptyl-4-hydroxyquinoline N-oxide, an antistaphylococcal agent produced by Pseudomonas aeruginosa. J. Antimicrob. Chemother. 30, 615–623 (1992).
Hoffman, L. R. et al. Selection for Staphylococcus aureus small-colony variants due to growth in the presence of Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA 103, 19890–19895 (2006).
Hotterbeekx, A., Kumar-Singh, S., Goossens, H. & Malhotra-Kumar, S. In vivo and in vitro interactions between Pseudomonas aeruginosa and Staphylococcus spp. Front. Cell Infect. Microbiol. 7, 106 (2017).
Chattoraj, S. S. et al. Rhinovirus infection liberates planktonic bacteria from biofilm and increases chemokine responses in cystic fibrosis airway epithelial cells. Thorax 66, 333–339 (2011).
Hendricks, M. R. et al. Respiratory syncytial virus infection enhances Pseudomonas aeruginosa biofilm growth through dysregulation of nutritional immunity. Proc. Natl Acad. Sci. USA 113, 1642–1647 (2016).
Kiedrowski, M. R. et al. Staphylococcus aureus biofilm growth on cystic fibrosis airway epithelial cells is enhanced during respiratory syncytial virus coinfection. mSphere 3, e00341-18 (2018).
Landi, A. et al. Pseudomonas aeruginosa lectin LecB impairs keratinocyte fitness by abrogating growth factor signalling. Life Sci. Alliance 2, e201900422 (2019).
Jordana-Lluch, E. et al. A simple polymicrobial biofilm keratinocyte colonization model for exploring interactions between commensals, pathogens and antimicrobials. Front. Microbiol. 11, 291 (2020).
Barrila, J. et al. Modeling host–pathogen interactions in the context of the microenvironment: three-dimensional cell culture comes of age. Infect. Immun. 86, e00282-18 (2018).
Bartfeld, S. Modeling infectious diseases and host-microbe interactions in gastrointestinal organoids. Dev. Biol. 420, 262–270 (2016).
Sachs, N. et al. Long-term expanding human airway organoids for disease modeling. EMBO J. 38, e100300 (2019).
Edwards, S. & Kjellerup, B. V. Exploring the applications of invertebrate host–pathogen models for in vivo biofilm infections. FEMS Immunol. Med. Microbiol. 65, 205–214 (2012).
Wiles, T. J. et al. Host gut motility promotes competitive exclusion within a model intestinal microbiota. PLoS Biol. 14, e1002517 (2016).
Bergeron, A. C. et al. Candida albicans and Pseudomonas aeruginosa interact to enhance virulence of mucosal infection in transparent zebrafish. Infect. Immun. 85, e00475-17 (2017).
Mulcahy, H., Sibley, C. D., Surette, M. G. & Lewenza, S. Drosophila melanogaster as an animal model for the study of Pseudomonas aeruginosa biofilm infections in vivo. PLoS Pathog. 7, e1002299 (2011).
Limmer, S. et al. Pseudomonas aeruginosa RhlR is required to neutralize the cellular immune response in a Drosophila melanogaster oral infection model. Proc. Natl Acad. Sci. USA 108, 17378–17383 (2011).
Garsin, D. A. et al. A simple model host for identifying Gram-positive virulence factors. Proc. Natl Acad. Sci. USA 98, 10892–10897 (2001).
Begun, J. et al. Staphylococcal biofilm exopolysaccharide protects against Caenorhabditis elegans immune defenses. PLoS Pathog. 3, e57 (2007).
Rezzoagli, C., Granato, E. T. & Kummerli, R. In-vivo microscopy reveals the impact of Pseudomonas aeruginosa social interactions on host colonization. ISME J. 13, 2403–2414 (2019).
Chua, S. L. et al. Dispersed cells represent a distinct stage in the transition from bacterial biofilm to planktonic lifestyles. Nat. Commun. 5, 4462 (2014).
Cassat, J. E. et al. Integrated molecular imaging reveals tissue heterogeneity driving host-pathogen interactions. Sci. Transl Med. 10, eaan6361 (2018).
Perry, W. J. et al. Staphylococcus aureus exhibits heterogeneous siderophore production within the vertebrate host. Proc. Natl Acad. Sci. USA 116, 21980–21982 (2019).
Tropini, C., Earle, K. A., Huang, K. C. & Sonnenburg, J. L. The gut microbiome: connecting spatial organization to function. Cell Host Microbe 21, 433–442 (2017).
Nava, G. M., Friedrichsen, H. J. & Stappenbeck, T. S. Spatial organization of intestinal microbiota in the mouse ascending colon. ISME J. 5, 627–638 (2011).
Keilberg, D., Zavros, Y., Shepherd, B., Salama, N. R. & Ottemann, K. M. Spatial and temporal shifts in bacterial biogeography and gland occupation during the development of a chronic infection. mBio 7, e01705-16 (2016).
Mark Welch, J. L., Hasegawa, Y., McNulty, N. P., Gordon, J. I. & Borisy, G. G. Spatial organization of a model 15-member human gut microbiota established in gnotobiotic mice. Proc. Natl Acad. Sci. USA 114, E9105–E9114 (2017).
Harrison, F. & Diggle, S. P. An ex vivo lung model to study bronchioles infected with Pseudomonas aeruginosa biofilms. Microbiology 162, 1755–1760 (2016).
Harrison, F., Muruli, A., Higgins, S. & Diggle, S. P. Development of an ex vivo porcine lung model for studying growth, virulence, and signaling of Pseudomonas aeruginosa. Infect. Immun. 82, 3312–3323 (2014).
Sweeney, E. et al. An ex vivo cystic fibrosis model recapitulates key clinical aspects of chronic Staphylococcus aureus infection. Microbiology 167, 000987 (2021).
Hassan, M. M., Harrington, N. E., Sweeney, E. & Harrison, F. Predicting antibiotic-associated virulence of Pseudomonas aeruginosa using an ex vivo lung biofilm model. Front. Microbiol. 11, 568510 (2020).
Cheong, J. Z. A. et al. Priority effects dictate community structure and alter virulence of fungal–bacterial biofilms. ISME J. 17, 2012-2027 (2021).
Jorth, P., Spero, M. A., Livingston, J. & Newman, D. K. Quantitative visualization of gene expression in mucoid and nonmucoid Pseudomonas aeruginosa aggregates reveals localized peak expression of alginate in the hypoxic zone. mBio 10, e02622-19 (2019).
Dar, D., Dar, N., Cai, L. & Newman, D. K. Spatial transcriptomics of planktonic and sessile bacterial populations at single-cell resolution. Science 373, eabi4882 (2021). This study assesses the gene expression heterogeneity in spatially organized aggregates of P. aeruginosa at the single-cell level by developing the par-seqFISH technique.
Wang, X. et al. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 361, eaat5691 (2018). This study develops and uses the single-cell approach STARmap to determine changes in the transcriptional profile across space.
Shah, S. et al. Single-molecule RNA detection at depth by hybridization chain reaction and tissue hydrogel embedding and clearing. Development 143, 2862–2867 (2016).
Stahl, P. L. et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82 (2016).
Järbrink, K. et al. The humanistic and economic burden of chronic wounds: a protocol for a systematic review. Syst. Rev. 6, 15 (2017).
Connell, J. L. et al. Probing prokaryotic social behaviors with bacterial “lobster traps”. mBio 1, e00202-10 (2010).
Heydorn, A. et al. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146, 2395–2407 (2000).
Nilsson, M., Givskov, M., Twetman, S. & Tolker-Nielsen, T. Inactivation of the pgmA gene in Streptococcus mutans significantly decreases biofilm-associated antimicrobial tolerance. Microorganisms 7, 310 (2019).
Beaudoin, T. et al. Staphylococcus aureus interaction with Pseudomonas aeruginosa biofilm enhances tobramycin resistance. npj Biofilms Microbiomes 3, 25 (2017).
Ciofu, O., Mandsberg, L. F., Wang, H. & Hoiby, N. Phenotypes selected during chronic lung infection in cystic fibrosis patients: implications for the treatment of Pseudomonas aeruginosa biofilm infections. FEMS Immunol. Med. Microbiol. 65, 215–225 (2012).
Daims, H., Lucker, S. & Wagner, M. daime, a novel image analysis program for microbial ecology and biofilm research. Env. Microbiol. 8, 200–213 (2006).
Hartmann, R. et al. Quantitative image analysis of microbial communities with BiofilmQ. Nat. Microbiol. 6, 151–156 (2021).
Acknowledgements
The authors thank the Cystic Fibrosis foundation for a postdoctoral fellowship to S.A. (AZIMI18F0); the National Institutes of Health (NIH) for funding to G.R.L. (F32DE027281 and K99DE031018); and NIH Grants R01DE023193, R01DE020100 and 1R01GM116547 and a grant from the Shurl and Kay Curci Foundation to M.W.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Microbiology thanks Gary Borisy, Kevin Foster, Nicholas Jakubovics and Jessica Mark Welch for their contribution to the peer review of this work.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Biogeography
-
The spatial assembly and distribution of various organisms in an environment through time.
- Microbiogeography
-
The spatial patterning of microorganisms at the micron scale within a single environment.
- Quorum sensing
-
The detection of an increased concentration of small signal molecules at high cell density, which can control gene expression in bacterial populations.
- Cystic fibrosis
-
A disorder that is caused by mutation(s) in the cystic fibrosis transmembrane conductance regulator gene, which affects cells that produce mucus, sweat and digestive enzymes. In people with cystic fibrosis, the build-up of mucus in the airways results in chronic polymicrobial lung infections.
- Function
-
A physiological property of a bacterium, such as glucose catabolism or motility.
- Catalysed reporter deposition FISH
-
(CARD-FISH). In CARD-FISH, the nucleotide probe is conjugated to horseradish peroxidase that amplifies the fluorescence in situ hybridization (FISH) signal. This method is used for bacterial taxa or bacterial functions with inherently low signal, for example bacteria with low ribosome content.
- Double labelling of oligonucleotide probes FISH
-
(DOPE-FISH). DOPE-FISH uses 5′ and 3′ double-labelled oligonucleotide probes to increase the fluorescence in situ hybridization (FISH) signal.
- Combinatorial labelling and spectral imaging FISH
-
(CLASI-FISH). Simultaneous fluorescence in situ hybridization (FISH) labelling of individual taxa using multiple fluorophores. By mixing the fluorophore combinations used for each taxon, this method enables the detection of tens to hundreds of taxa concurrently. Spectral imaging is usually followed by linear unmixing analysis to differentiate between taxa.
- Alginate
-
An exopolysaccharide produced by Pseudomonas aeruginosa that helps cells to attach to surfaces and form biofilms.
- Extracellular DNA
-
(eDNA). eDNA can be produced by host cells or bacteria. It is one of the main components of the biofilm matrix that can provide structural scaffold for bacterial cells and may play a role in protecting bacterial cells in response to host cells and antibiotics.
- NETosis
-
Neutrophil extracellular traps (NETs) are nucleic acid and intracellular components that are forced out of polymorphonuclear leukocytes in response to microorganisms and proinflammatory cytokines. NETosis is a type of programmed cell death in polymorphonuclear leukocytes such as neutrophils that functions as cellular defence. During NETosis, cells force out their chromatin, forming sticky traps that can trap bacterial cells.
- Mucin
-
A type of high molecular weight glycosylated protein produced by epithelial cells in animals that forms gels and is found in high levels in lung infections. Bovine and porcine mucins can be purchased commercially, which has led to their use in numerous preclinical models.
- Hybridization chain reaction
-
The detection and quantification of RNA transcripts using exogenously added fluorophore-conjugated DNA hairpins. These hairpins self-assemble, amplifying their signal.
- Coagulase
-
An enzyme produced by several types of bacteria that converts the soluble protein fibrinogen in blood to insoluble fibrin, leading to clot formation.
- Preclinical infection models
-
In vitro and in vivo models used to study microbial infection.
- Raman microscopy
-
A combination of laser-based imaging and Raman spectroscopy to detect the differential excitation levels of photons. This method is used to identify certain molecules without disturbing the spatial arrangement of the samples.
- Atomic force microscopy
-
A powerful imaging technique that uses a fine and specific tip attached to a cantilever that scans along the surface of a sample. The changes in contact forces between the tip and the surface of the sample are recorded by a laser beam that generates an accurate topographic image of the surface at nanometre resolution.
- Surface plasmon resonance imaging
-
A label-free imaging method that is used to detect and measure the attachment level and surface properties of bacterial biofilms. This method analyses changes in the angle of reflected light of a surface covered with the sample of interest, compared with a control surface.
- Scanning electrochemical microscopy
-
A label-free microscopy method using probes that detect redox reactions and can provide an electrochemical map of an environment.
- Imaging mass spectrometry
-
Mass spectrometry performed across a spatial plane to build a map of detected chemicals.
- Hydroxyapatite
-
A mineral form of calcium apatite that is the main component of tooth enamel and bones.
- Fibrin
-
A glycosylated protein in blood formed by the enzymatic action of a serine protease on soluble fibrinogen. Polymerized fibrin leads to clotting of the blood.
- Transferrin
-
A glycoprotein that binds to iron and transports iron through the bloodstream.
- Keratinocyte
-
A cell that forms the outer layer of skin and produces keratin to form a protective barrier.
- Organoid
-
A small, differentiated collection of cells containing similar cell types and functions as an organ. Organoids are produced in vitro using stem cells derived from the organ of interest that are cultured in a medium containing growth factors and extracellular matrix.
- Organ-on-a-chip
-
A cell culture device that contains a multichannel 3D microfluidics chip, designed to mimic the physical and chemical properties of an organ. Organ-on-a-chip models can provide a structured microenvironment for high-throughput assessment of bacterial–host interactions, for instance in response to various stimuli.
- Glycol methacrylate resin
-
An ester form of epoxy resin that can be used instead of paraffin for embedding biological samples for better quality imaging.
Rights and permissions
About this article
Cite this article
Azimi, S., Lewin, G.R. & Whiteley, M. The biogeography of infection revisited. Nat Rev Microbiol 20, 579–592 (2022). https://doi.org/10.1038/s41579-022-00683-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41579-022-00683-3
This article is cited by
-
Spatial transcriptome uncovers rich coordination of metabolism in E. coli K12 biofilm
Nature Chemical Biology (2023)
-
Selenomonas sputigena acts as a pathobiont mediating spatial structure and biofilm virulence in early childhood caries
Nature Communications (2023)
-
Advances in the screening of antimicrobial compounds using electrochemical biosensors: is there room for nanomaterials?
Analytical and Bioanalytical Chemistry (2023)
-
Dispersing biofilm myths
Nature Reviews Microbiology (2022)