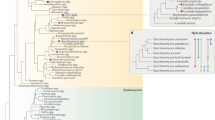
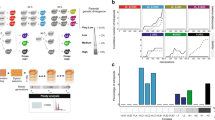
Abstract
Hybridization between species often leads to non-viable or infertile offspring, yet examples of evolutionarily successful interspecific hybrids have been reported in all kingdoms of life. However, many questions on the ecological circumstances and evolutionary aftermath of interspecific hybridization remain unanswered. In this study, we sequenced and phenotyped a large set of interspecific yeast hybrids isolated from brewing environments to uncover the influence of interspecific hybridization in yeast adaptation and domestication. Our analyses demonstrate that several hybrids between Saccharomyces species originated and diversified in industrial environments by combining key traits of each parental species. Furthermore, posthybridization evolution within each hybrid lineage reflects subspecialization and adaptation to specific beer styles, a process that was accompanied by extensive chimerization between subgenomes. Our results reveal how interspecific hybridization provides an important evolutionary route that allows swift adaptation to novel environments.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All raw sequencing reads generated in this study have been deposited to the NCBI Short Read Archive (BioProject accession: PRJNA516557).
References
Mallet, J., Besansky, N. & Hahn, M. W. How reticulated are species? BioEssays 38, 140–149 (2016).
Abbott, R. et al. Hybridization and speciation. J. Evol. Biol. 26, 229–246 (2013).
Shapiro, B. J., Leducq, J.-B. & Mallet, J. What is speciation? PLoS Genet. 12, e1005860 (2016).
Nolte, A. W. & Tautz, D. Understanding the onset of hybrid speciation. Trends Genet. 26, 54–58 (2010).
Baack, E. J. & Rieseberg, L. H. A genomic view of introgression and hybrid speciation. Curr. Opin. Genet. Dev. 17, 513–518 (2007).
Rieseberg, L. H., Archer, M. A. & Wayne, R. K. Transgressive segregation, adaptation and speciation. Heredity 83, 363–372 (1999).
Lamichhaney, S. et al. Rapid hybrid speciation in Darwin’s finches. Science 359, 224–228 (2018).
Rieseberg, L. H. et al. Hybridization and the colonization of novel habitats by annual sunflowers. Genetica 129, 149–165 (2007).
Rieseberg, L. H. Homoploid reticulate evolution in Helianthus (Asteraceae): evidence from ribosomal genes. Am. J. Bot. 78, 1218–1237 (1991).
Schwarzbach, A. E. & Rieseberg, L. H. Likely multiple origins of a diploid hybrid sunflower species. Mol. Ecol. 11, 1703–1715 (2002).
Pryszcz, L. P. et al. The genomic aftermath of hybridization in the opportunistic pathogen Candida metapsilosis. PLoS Genet. 11, e1005626 (2015).
Kellis, M., Patterson, N., Endrizzi, M., Birren, B. & Lander, E. S. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423, 241–254 (2003).
Greig, D. Reproductive isolation in Saccharomyces. Heredity 102, 39–44 (2009).
Liti, G., Barton, D. B. H. & Louis, E. J. Sequence diversity, reproductive isolation and species concepts in Saccharomyces. Genetics 174, 839–850 (2006).
Hou, J., Friedrich, A., De Montigny, J. & Schacherer, J. Chromosomal rearrangements as a major mechanism in the onset of reproductive isolation in Saccharomyces cerevisiae. Curr. Biol. 24, 1153–1159 (2014).
Morales, L. & Dujon, B. Evolutionary role of interspecies hybridization and genetic exchanges in yeasts. Microbiol. Mol. Biol. Rev. 76, 721–739 (2012).
Naumov, G. I. et al. Natural polyploidization of some cultured yeast Saccharomyces sensu stricto: auto- and allotetraploidy. Syst. Appl. Microbiol. 23, 442–449 (2000).
Zhang, H., Skelton, A., Gardner, R. C. & Goddard, M. R. Saccharomyces paradoxus and Saccharomyces cerevisiae reside on oak trees in New Zealand: evidence for migration from Europe and interspecies hybrids. FEMS Yeast Res. 10, 941–947 (2010).
Stefanini, I. et al. Social wasps are a Saccharomyces mating nest. Proc. Natl Acad. Sci. USA 113, 2247–2251 (2016).
Nakao, Y. et al. Genome sequence of the lager brewing yeast, an interspecies hybrid. DNA Res. 16, 115–129 (2009).
Walther, A., Hesselbart, A. & Wendland, J. Genome sequence of Saccharomyces carlsbergensis, the world’s first pure culture lager yeast. G3 4, 783–793 (2014).
Dunn, B. & Sherlock, G. Reconstruction of the genome origins and evolution of the hybrid lager yeast Saccharomyces pastorianus. Genome Res. 18, 1610–1623 (2008).
Bing, J., Han, P.-J., Liu, W.-Q., Wang, Q.-M. & Bai, F.-Y. Evidence for a Far East Asian origin of lager beer yeast. Curr. Biol. 24, R380–R381 (2014).
Cavalieri, D., McGovern, P. E., Hartl, D. L., Mortimer, R. & Polsinelli, M. Evidence for S. cerevisiae fermentation in ancient wine. J. Mol. Evol. 57 S226–S232 (2003).
Michel, R. H., McGovern, P. E. & Badler, V. R. Chemical evidence for ancient beer. Nature 360, 24 (1992).
McGovern, P. E. et al. Fermented beverages of pre- and proto-historic china. Proc. Natl Acad. Sci. USA 101, 17593–17598 (2004).
Borneman, A. R. et al. Whole-genome comparison reveals novel genetic elements that characterize the genome of industrial strains of Saccharomyces cerevisiae. PLoS Genet. 7, e1001287 (2011).
Gallone, B. et al. Domestication and divergence of Saccharomyces cerevisiae beer yeasts. Cell 166, 1397–1410.e16 (2016).
Gonçalves, M. et al. Distinct domestication trajectories in top-fermenting beer yeasts and wine yeasts. Curr. Biol. 26, 2750–2761 (2016).
Legras, J.-L. et al. Adaptation of S. cerevisiae to fermented food environments reveals remarkable genome plasticity and the footprints of domestication. Mol. Biol. Evol. 35, 1712–1727 (2018).
Peter, J. et al. Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature 556, 339–344 (2018).
Yue, J.-X. et al. Contrasting evolutionary genome dynamics between domesticated and wild yeasts. Nat. Genet. 49, 913–924 (2017).
Dunn, B., Richter, C., Kvitek, D. J., Pugh, T. & Sherlock, G. Analysis of the Saccharomyces cerevisiae pan-genome reveals a pool of copy number variants distributed in diverse yeast strains from differing industrial environments. Genome Res. 22, 908–924 (2012).
Duan, S. F. et al. The origin and adaptive evolution of domesticated populations of yeast from Far East Asia. Nat. Commun. 9, 2690 (2018).
Ludlow, C. L. et al. Independent origins of yeast associated with coffee and cacao fermentation. Curr. Biol. 26, 965–971 (2016).
Barbosa, R. et al. Multiple rounds of artificial selection promote microbe secondary domestication—the case of cachaça yeasts. Genome Biol. Evol. 10, 1939–1955 (2018).
Libkind, D. et al. Microbe domestication and the identification of the wild genetic stock of lager-brewing yeast. Proc. Natl Acad. Sci. USA 108, 14539–14544 (2011).
Eizaguirre, J. I. et al. Phylogeography of the wild lager-brewing ancestor (Saccharomyces eubayanus) in Patagonia. Environ. Microbiol. 20, 3732–3743 (2018).
Gibson, B. R., Storgårds, E., Krogerus, K. & Vidgren, V. Comparative physiology and fermentation performance of Saaz and Frohberg lager yeast strains and the parental species Saccharomyces eubayanus. Yeast 30, 255–266 (2013).
González, S. S., Barrio, E. & Querol, A. Molecular characterization of new natural hybrids of Saccharomyces cerevisiae and S. kudriavzevii in brewing. Appl. Environ. Microbiol. 74, 2314–2320 (2008).
González, S. S., Barrio, E., Gafner, J. & Querol, A. Natural hybrids from Saccharomyces cerevisiae, Saccharomyces bayanus and Saccharomyces kudriavzevii in wine fermentations. FEMS Yeast Res. 6, 1221–1234 (2006).
Masneuf, I., Hansen, J., Groth, C., Piskur, J. & Dubourdieu, D. New hybrids between Saccharomyces sensu stricto yeast species found among wine and cider production strains. Appl. Environ. Microbiol. 64, 3887–3892 (1998).
Peris, D., Lopes, Ca, Belloch, C., Querol, A. & Barrio, E. Comparative genomics among Saccharomyces cerevisiae × Saccharomyces kudriavzevii natural hybrid strains isolated from wine and beer reveals different origins. BMC Genomics 13, 407 (2012).
Peris, D., Pérez-Torrado, R., Hittinger, C. T., Barrio, E. & Querol, A. On the origins and industrial applications of Saccharomyces cerevisiae × Saccharomyces kudriavzevii hybrids. Yeast 35, 51–69 (2018).
Lopes, C. A., Barrio, E. & Querol, A. Natural hybrids of S. cerevisiae × S. kudriavzevii share alleles with European wild populations of Saccharomyces kudriavzevii. FEMS Yeast Res. 10, 412–421 (2010).
Peris, D. et al. The molecular characterization of new types of Saccharomyces cerevisiae × S. kudriavzevii hybrid yeasts unveils a high genetic diversity. Yeast 29, 81–91 (2012).
Le Jeune, C. et al. Characterization of natural hybrids of Saccharomyces cerevisiae and Saccharomyces bayanus var. uvarum. FEMS Yeast Res. 7, 540–549 (2007).
Gallone, B. et al. Origins, evolution, domestication and diversity of Saccharomyces beer yeasts. Curr. Opin. Biotechnol. 49, 148–155 (2018).
Steensels, J., Gallone, B., Voordeckers, K. & Verstrepen, K. J. Domestication of industrial microbes. Curr. Biol. 29, R381–R393 (2019).
Wendland, J. Lager yeast comes of age. Eukaryot. Cell 13, 1256–1265 (2014).
G Gibson, B. & Liti, G. Saccharomyces pastorianus: genomic insights inspiring innovation for industry. Yeast 32, 17–27 (2015).
Peris, D. et al. Complex ancestries of lager-brewing hybrids were shaped by standing variation in the wild yeast Saccharomyces eubayanus. PLoS Genet. 12, e1006155 (2016).
Okuno, M. et al. Next-generation sequencing analysis of lager brewing yeast strains reveals the evolutionary history of interspecies hybridization. DNA Res. 23, dsv037 (2016).
Dai, F. et al. Tibet is one of the centers of domestication of cultivated barley. Proc. Natl Acad. Sci. USA 109, 16969–16973 (2012).
Meußdoerffer, F. & Zarnkow, M. Das Bier: Eine Geschichte von Hopfen und Malz (Beck, 2014).
Baker, E. et al. The genome sequence of Saccharomyces eubayanus and the domestication of lager-brewing yeasts. Mol. Biol. Evol. 32, 2818–2831 (2015).
Monerawela, C. & Bond, U. Recombination sites on hybrid chromosomes in Saccharomyces pastorianus share common sequence motifs and define a complex evolutionary relationship between group I and II lager yeasts. FEMS Yeast Res. 17, fox047 (2017).
Belloch, C. et al. Chimeric genomes of natural hybrids of Saccharomyces cerevisiae and Saccharomyces kudriavzevii. Appl. Environ. Microbiol. 75, 2534–2544 (2009).
Hewitt, S. K., Donaldson, I. J., Lovell, S. C. & Delneri, D. Sequencing and characterisation of rearrangements in three S. pastorianus strains reveals the presence of chimeric genes and gives evidence of breakpoint reuse. PLoS ONE 9, e92203 (2014).
Almeida, P. et al. A Gondwanan imprint on global diversity and domestication of wine and cider yeast Saccharomyces uvarum. Nat. Commun. 5, 4044 (2014).
Peŕez-Través, L., Lopes, C. A., Querol, A. & Barrio, E. On the complexity of the Saccharomyces bayanus taxon: hybridization and potential hybrid speciation. PLoS ONE 9, e93729 (2014).
Back, W. Colour Atlas and Handbook of Beverage Biology (Fachverlag Hans Carl, 2005).
Krogerus, K., Magalhães, F., Vidgren, V. & Gibson, B. Novel brewing yeast hybrids: creation and application. Appl. Microbiol. Biotechnol. 101, 65–78 (2017).
van den Broek, M. et al. Chromosomal copy number variation in Saccharomyces pastorianus is evidence for extensive genome dynamics in industrial lager brewing strains. Appl. Environ. Microbiol. 81, 6253–6267 (2015).
Monerawela, C. & Bond, U. The hybrid genomes of Saccharomyces pastorianus: a current perspective. Yeast 35, 39–50 (2017).
Fisher, K. J., Buskirk, S. W., Vignogna, R. C., Marad, D. A. & Lang, G. I. Adaptive genome duplication affects patterns of molecular evolution in Saccharomyces cerevisiae. PLoS Genet. 14, e1007396 (2018).
Selmecki, A. M. et al. Polyploidy can drive rapid adaptation in yeast. Nature 519, 349–352 (2015).
Todd, R. T., Forche, A. & Selmecki, A. in The Fungal Kingdom (eds Heitman, J. et al.) Ch. 28 (ASM Press, 2017).
Baker, E. C. P. et al. Mitochondrial DNA and temperature tolerance in lager yeasts. Sci. Adv. 5, eaav1869 (2019).
Li, X. C., Peris, D., Hittinger, C. T., Sia, E. A. & Fay, J. C. Mitochondria-encoded genes contribute to evolution of heat and cold tolerance in yeast. Sci. Adv. 5, eaav1848 (2019).
De Roos, J. & De Vuyst, L. Microbial acidification, alcoholization, and aroma production during spontaneous lambic beer production. J. Sci. Food Agric. 99, 25–38 (2019).
Steensels, J. & Verstrepen, K. J. Taming wild yeast: potential of conventional and nonconventional yeasts in industrial fermentations. Annu. Rev. Microbiol. 68, 61–80 (2014).
Dzialo, M. C., Park, R., Steensels, J., Lievens, B. & Verstrepen, K. J. Physiology, ecology and industrial applications of aroma formation in yeast. FEMS Microbiol. Rev. 41, S95–S128 (2017).
Mukai, N., Masaki, K., Fujii, T., Kawamukai, M. & Iefuji, H. PAD1 and FDC1 are essential for the decarboxylation of phenylacrylic acids in Saccharomyces cerevisiae. J. Biosci. Bioeng. 109, 564–569 (2010).
Mukai, N., Masaki, K., Fujii, T. & Iefuji, H. Single nucleotide polymorphisms of PAD1 and FDC1 show a positive relationship with ferulic acid decarboxylation ability among industrial yeasts used in alcoholic beverage production. J. Biosci. Bioeng. 118, 50–55 (2014).
Pasteur, L. Etudes sur la bière, ses maladies, causes qui les provoquent, procédé pour la rendre inaltérable, avec une théorie nouvelle de la fermentation (Gauthier-Villars, 1876).
Teich, M. Bier, Wissenschaft und Wirtschaft in Deutschland 1800–1914. Ein Beitrag zur Deutschen Industrialisierungsgeschichte (Böhlau, 2000).
Hansen, E. Recherches sur la physiologie et la morphologie des ferments alcooliques V. methodes pour obtenir des cultures pures de Saccharomyces et de mikroorganismes analogues. C. R. Trav. Lab. Carlsberg 2, 92–105 (1883).
Mertens, S. et al. A large set of newly created interspecific Saccharomyces hybrids increases aromatic diversity in lager beers. Appl. Environ. Microbiol. 81, 8202–8214 (2015).
Peris, D. et al. Hybridization and adaptive evolution of diverse Saccharomyces species for cellulosic biofuel production. Biotechnol. Biofuels 10, 78 (2017).
Scannell, D. R. et al. The awesome power of yeast evolutionary genetics: new genome sequences and strain resources for the Saccharomyces sensu stricto genus. G3 1, 11–25 (2011).
Liti, G. et al. High quality de novo sequencing and assembly of the Saccharomyces arboricolus genome. BMC Genomics 14, 69 (2013).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Bankevich, A. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012).
Pryszcz, L. P. & Gabaldón, T. Redundans: an assembly pipeline for highly heterozygous genomes. Nucleic Acids Res. 44, e113 (2016).
Kolmogorov, M., Raney, B., Paten, B. & Pham, S. Ragout—a reference-assisted assembly tool for bacterial genomes. Bioinformatics 30, i302–i309 (2014).
Wapinski, I., Pfeffer, A., Friedman, N. & Regev, A. Natural history and evolutionary principles of gene duplication in fungi. Nature 449, 54–61 (2007).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Camacho, C. et al. BLAST+: architecture and applications. BMC Bioinformatics 10, 421 (2009).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Kück, P. & Meusemann, K. FASconCAT: convenient handling of data matrices. Mol. Phylogenet. Evol. 56, 1115–1118 (2010).
Capella-Gutiérrez, S., Silla-Martínez, J. M. & Gabaldón, T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Kozlov, A. M., Aberer, A. J. & Stamatakis, A. ExaML version 3: a tool for phylogenomic analyses on supercomputers. Bioinformatics 31, 2577–2579 (2015).
Pattengale, N. D., Alipour, M., Bininda-Emonds, O. R. P., Moret, B. M. E. & Stamatakis, A. How many bootstrap replicates are necessary? J. Comput. Biol. 17, 337–354 (2010).
R Core Team R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2011).
Yu, G., Smith, D. K., Zhu, H., Guan, Y. & Lam, T. T. Y. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8, 28–36 (2017).
Suchard, M. A. et al. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 4, vey016 (2018).
Raj, A., Stephens, M. & Pritchard, J. K. fastSTRUCTURE: variational inference of population structure in large SNP data sets. Genetics 197, 573–589 (2014).
Ayres, D. L. et al. BEAGLE: an application programming interface and high-performance computing library for statistical phylogenetics. Syst. Biol. 61, 170–173 (2012).
Drummond, A. J., Ho, S. Y., Phillips, M. J. & Rambaut, A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4, e88 (2006).
Drummond, A. J. & Suchard, M. A. Bayesian random local clocks, or one rate to rule them all. BMC Biol. 8, 114 (2010).
Worobey, M., Han, G.-Z. & Rambaut, A. A synchronized global sweep of the internal genes of modern avian influenza virus. Nature 508, 254–257 (2014).
Rambaut, A., Drummond, A. J., Xie, D., Baele, G. & Suchard, M. A. Posterior summarization in Bayesian phylogenetics using tracer 1.7. Syst. Biol. 67, 901–904 (2018).
Baele, G., Lemey, P. & Suchard, M. A. Genealogical working distributions for Bayesian model testing with phylogenetic uncertainty. Syst. Biol. 65, 250–264 (2016).
Angiuoli, S. V. & Salzberg, S. L. Mugsy: fast multiple alignment of closely related whole genomes. Bioinformatics 27, 334–342 (2011).
Venables, W. N. & Ripley, B. D. Modern applied statistics with S. Technometrics 45, 111–111 (2003).
Warnes, G. R. et al. gplots: Various R programming tools for plotting data. R version 3.0.0 http://CRAN.R-project.org/package=gplots (2016).
Murtagh, F. & Legendre, P. Ward’s hierarchical agglomerative clustering method: which algorithms implement Ward’s criterion? J. Classif. 31, 274–295 (2014).
Calahan, D., Dunham, M., Desevo, C. & Koshland, D. E. Genetic analysis of desiccation tolerance in Saccharomyces cerevisiae. Genetics 189, 507–519 (2011).
Muir, A., Harrison, E. & Wheals, A. A multiplex set of species-specific primers for rapid identification of members of the genus Saccharomyces. FEMS Yeast Res. 11, 552–563 (2011).
Pengelly, R. J. & Wheals, A. E. Rapid identification of Saccharomyces eubayanus and its hybrids. FEMS Yeast Res. 13, 156–161 (2013).
Acknowledgements
We thank all K.J.V. and S.Maere laboratory members for their help and suggestions. We thank AB InBev, White Labs Inc. and R. S. Wagner (Central Washington University) for isolating and sharing various hybrid yeasts from the beer environment. We thank M. Zarnkow (Research Center Weihenstephan for Brewing and Food Quality) for sharing historical facts on lager brewing. B.G. acknowledges funding from the VIB International PhD Program in Life Sciences. J.S. acknowledges funding from Research Foundation-Flanders (FWO) (grant no. 12W3918N). Research in the laboratory of K.J.V. is supported by KU Leuven Program Financing, European Research Council Consolidator Grant (no. CoG682009), Human Frontier Science program grant (no. RGP0050/2013), VIB, FWO (grant no. G.0C59.14N) and Vlaams Agentschap Innoveren en Ondernemen (grant no. HBC.2018.0097). Research in the laboratory of S.Maere is supported by VIB, Ghent University and FWO (grant no. G018915N).
Author information
Authors and Affiliations
Contributions
B.G., J.S., S.Maere and K.J.V. conceived the project. B.G. and J.S. performed the data collection. B.G., J.S. and G.B. carried out the formal analysis. B.G., J.S., S.Mertens, J.L.G., R.W., F.A.T., F.B., V.S., B.H.-M. and G.B. carried out the investigations. B.G. and J.S. performed data visualization. M.H., F.M., P.M., B.S., L.D., T.P., C.W. and K.J.V. provided resources. B.G., J.S., M.C.D., S.Maere and K.J.V. wrote the paper. S.Maere and K.J.V. supervised the project.
Corresponding authors
Ethics declarations
Competing interests
AB InBev partially funded the experiments performed in this study. AB InBev had no role in conceptualization, design, data collection, analysis, decision to publish or preparation of the manuscript. B.S., P.M. and L.D. are employed by AB InBev.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–5, Tables 1–3, Note 1 and Dataset 1.
Supplementary Table 1
List of strains included in this study.
Supplementary Table 2
Chimeric breakpoints identified in Saccharomyces interspecific hybrids.
Supplementary Table 3
Phenotypic variation of Saccharomyces interspecific hybrids and Saccharomyces pure species.
Rights and permissions
About this article
Cite this article
Gallone, B., Steensels, J., Mertens, S. et al. Interspecific hybridization facilitates niche adaptation in beer yeast. Nat Ecol Evol 3, 1562–1575 (2019). https://doi.org/10.1038/s41559-019-0997-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41559-019-0997-9
This article is cited by
-
Ancient and recent origins of shared polymorphisms in yeast
Nature Ecology & Evolution (2024)
-
phyBWT2: phylogeny reconstruction via eBWT positional clustering
Algorithms for Molecular Biology (2023)
-
Increased volatile thiol release during beer fermentation using constructed interspecies yeast hybrids
European Food Research and Technology (2023)
-
Macroevolutionary diversity of traits and genomes in the model yeast genus Saccharomyces
Nature Communications (2023)
-
Unlocking the functional potential of polyploid yeasts
Nature Communications (2022)