Abstract
Following severe or chronic liver injury, adult ductal cells (cholangiocytes) contribute to regeneration by restoring both hepatocytes and cholangiocytes. We recently showed that ductal cells clonally expand as self-renewing liver organoids that retain their differentiation capacity into both hepatocytes and ductal cells. However, the molecular mechanisms by which adult ductal-committed cells acquire cellular plasticity, initiate organoids and regenerate the damaged tissue remain largely unknown. Here, we describe that ductal cells undergo a transient, genome-wide, remodelling of their transcriptome and epigenome during organoid initiation and in vivo following tissue damage. TET1-mediated hydroxymethylation licences differentiated ductal cells to initiate organoids and activate the regenerative programme through the transcriptional regulation of stem-cell genes and regenerative pathways including the YAP–Hippo signalling. Our results argue in favour of the remodelling of genomic methylome/hydroxymethylome landscapes as a general mechanism by which differentiated cells exit a committed state in response to tissue damage.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The RNA, ChIP, DamID, WGBS and RRHP sequencing data that support the findings of this study have been deposited in the Gene Expression Omnibus under the accession code GSE123133.
All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Code availability
All codes used are available on request.
References
Choi, T. Y., Ninov, N., Stainier, D. Y. & Shin, D. Extensive conversion of hepatic biliary epithelial cells to hepatocytes after near total loss of hepatocytes in zebrafish. Gastroenterology 146, 776–788 (2014).
Russell, J. O. et al. Hepatocyte-specific β-catenin deletion during severe liver injury provokes cholangiocytes to differentiate into hepatocytes. Hepatology 69, 742–759 (2019).
Espanol-Suner, R. et al. Liver progenitor cells yield functional hepatocytes in response to chronic liver injury in mice. Gastroenterology 143, 1564–1575 (2012).
Huch, M. et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature 494, 247–250 (2013).
Lu, W. Y. et al. Hepatic progenitor cells of biliary origin with liver repopulation capacity. Nat. Cell Biol. 17, 971–983 (2015).
Sackett, S. D. et al. Foxl1 is a marker of bipotential hepatic progenitor cells in mice. Hepatology 49, 920–929 (2009).
Shin, S. et al. Foxl1-Cre-marked adult hepatic progenitors have clonogenic and bilineage differentiation potential. Genes Dev. 25, 1185–1192 (2011).
Deng, X. et al. Chronic liver injury induces conversion of biliary epithelial cells into hepatocytes. Cell Stem Cell 23, 114–122 (2018).
Raven, A. et al. Cholangiocytes act as facultative liver stem cells during impaired hepatocyte regeneration. Nature 547, 350–354 (2017).
Barker, N. et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007).
Prior, N. et al. Lgr5+ stem and progenitor cells reside at the apex of a heterogeneous embryonic hepatoblast pool. Development 146, dev174557 (2019).
Okabe, M. et al. Potential hepatic stem cells reside in EpCAM+ cells of normal and injured mouse liver. Development 136, 1951–1960 (2009).
Huch, M. et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 160, 299–312 (2015).
Li, B. et al. Adult mouse liver contains two distinct populations of cholangiocytes. Stem Cell Reports 9, 478–489 (2017).
Messerschmidt, D. M., Knowles, B. B. & Solter, D. DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes Dev. 28, 812–828 (2014).
Iurlaro, M., von Meyenn, F. & Reik, W. DNA methylation homeostasis in human and mouse development. Curr. Opin. Genet. Dev. 43, 101–109 (2017).
Smith, Z. D. & Meissner, A. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 14, 204–220 (2013).
Bird, A. DNA methylation patterns and epigenetic memory. Genes Dev. 16, 6–21 (2002).
Li, E. & Zhang, Y. DNA methylation in mammals. Cold Spring Harb. Perspect. Biol. 6, a019133 (2014).
Probst, A. V., Dunleavy, E. & Almouzni, G. Epigenetic inheritance during the cell cycle. Nat. Rev. Mol. Cell Biol. 10, 192–206 (2009).
Kohli, R. M. & Zhang, Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature 502, 472–479 (2013).
Pastor, W. A., Aravind, L. & Rao, A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat. Rev. Mol. Cell Biol. 14, 341–356 (2013).
Hill, P. W., Amouroux, R. & Hajkova, P. DNA demethylation, Tet proteins and 5-hydroxymethylcytosine in epigenetic reprogramming: an emerging complex story. Genomics 104, 324–333 (2014).
Hahn, M. A., Szabo, P. E. & Pfeifer, G. P. 5-Hydroxymethylcytosine: a stable or transient DNA modification? Genomics 104, 314–323 (2014).
Branco, M. R., Ficz, G. & Reik, W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat. Rev. Genet. 13, 7–13 (2011).
Yamaguchi, S., Shen, L., Liu, Y., Sendler, D. & Zhang, Y. Role of Tet1 in erasure of genomic imprinting. Nature 504, 460–464 (2013).
Hill, P. W. S. et al. Epigenetic reprogramming enables the transition from primordial germ cell to gonocyte. Nature 555, 392–396 (2018).
Ficz, G. et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature 473, 398–402 (2011).
Costa, Y. et al. NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature 495, 370–374 (2013).
Rasmussen, K. D. & Helin, K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 30, 733–750 (2016).
Kim, R., Sheaffer, K. L., Choi, I., Won, K. J. & Kaestner, K. H. Epigenetic regulation of intestinal stem cells by Tet1-mediated DNA hydroxymethylation. Genes Dev. 30, 2433–2442 (2016).
Reizel, Y. et al. Postnatal DNA demethylation and its role in tissue maturation. Nat. Commun. 9, 2040 (2018).
Tarlow, B. D., Finegold, M. J. & Grompe, M. Clonal tracing of Sox9+ liver progenitors in mouse oval cell injury. Hepatology 60, 278–289 (2014).
Dorrell, C. et al. Prospective isolation of a bipotential clonogenic liver progenitor cell in adult mice. Genes Dev. 25, 1193–1203 (2011).
Mort, R. L. et al. Fucci2a: a bicistronic cell cycle reporter that allows Cre mediated tissue specific expression in mice. Cell Cycle 13, 2681–2696 (2014).
Duncan, A. W., Dorrell, C. & Grompe, M. Stem cells and liver regeneration. Gastroenterology 137, 466–481 (2009).
Medvedeva, Y. A. et al. EpiFactors: a comprehensive database of human epigenetic factors and complexes. Database 2015, bav067 (2015).
Huch, M. & Koo, B. K. Modeling mouse and human development using organoid cultures. Development 142, 3113–3125 (2015).
Tahiliani, M. et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 (2009).
Natarajan, A., Wagner, B. & Sibilia, M. The EGF receptor is required for efficient liver regeneration. Proc. Natl Acad. Sci. USA 104, 17081–17086 (2007).
Yang, L. et al. A single-cell transcriptomic analysis reveals precise pathways and regulatory mechanisms underlying hepatoblast differentiation. Hepatology 66, 1387–1401 (2017).
Marshall, O. J., Southall, T. D., Cheetham, S. W. & Brand, A. H. Cell-type-specific profiling of protein–DNA interactions without cell isolation using targeted DamID with next-generation sequencing. Nat. Protoc. 11, 1586–1598 (2016).
Cheetham, S. W. et al. Targeted DamID reveals differential binding of mammalian pluripotency factors. Development 145, dev.170209 (2018).
Hu, M. et al. Wnt/β-catenin signaling in murine hepatic transit amplifying progenitor cells. Gastroenterology 133, 1579–1591 (2007).
Wang, B., Zhao, L., Fish, M., Logan, C. Y. & Nusse, R. Self-renewing diploid Axin2+ cells fuel homeostatic renewal of the liver. Nature 524, 180–185 (2015).
Jensen, K. B. et al. Lrig1 expression defines a distinct multipotent stem cell population in mammalian epidermis. Cell Stem Cell 4, 427–439 (2009).
Chrysanthou, S. et al. A critical role of TET1/2 proteins in cell-cycle progression of trophoblast stem cells. Stem Cell Rep. 10, 1355–1368 (2018).
Boj, S. F. et al. Diabetes risk gene and Wnt effector Tcf7l2/TCF4 controls hepatic response to perinatal and adult metabolic demand. Cell 151, 1595–1607 (2012).
Fouraschen, S. M. et al. mTOR signaling in liver regeneration: rapamycin combined with growth factor treatment. World J. Transplant. 3, 36–47 (2013).
Planas-Paz, L. et al. YAP, but not RSPO-LGR4/5, signaling in biliary epithelial cells promotes a ductular reaction in response to liver injury. Cell Stem Cell 25, 39–53 (2019).
Talarmin, H. et al. The mitogen-activated protein kinase kinase/extracellular signal-regulated kinase cascade activation is a key signalling pathway involved in the regulation of G1 phase progression in proliferating hepatocytes. Mol. Cell. Biol. 19, 6003–6011 (1999).
Pepe-Mooney, B. J. et al. Single-cell analysis of the liver epithelium reveals dynamic heterogeneity and an essential role for YAP in homeostasis and regeneration. Cell Stem Cell 25, 23–38 (2019).
Yimlamai, D. et al. Hippo pathway activity influences liver cell fate. Cell 157, 1324–1338 (2014).
Serra, D. et al. Self-organization and symmetry breaking in intestinal organoid development. Nature 569, 66–72 (2019).
Lin, Y. et al. HGF/R-spondin1 rescues liver dysfunction through the induction of Lgr5+ liver stem cells. Nat. Commun. 8, 1175 (2017).
Kamimoto, K. et al. Heterogeneity and stochastic growth regulation of biliary epithelial cells dictate dynamic epithelial tissue remodeling. eLife 5, e15034 (2016).
Zhu, L. et al. Multi-organ mapping of cancer risk. Cell 166, 1132–1146 (2016).
Blanpain, C. & Fuchs, E. Stem cell plasticity. Plasticity of epithelial stem cells in tissue regeneration. Science 344, 1242281 (2014).
Lin, S. et al. Distributed hepatocytes expressing telomerase repopulate the liver in homeostasis and injury. Nature 556, 244–248 (2018).
Font-Burgada, J. et al. Hybrid periportal hepatocytes regenerate the injured liver without giving rise to cancer. Cell 162, 766–779 (2015).
Huch, M. & Dolle, L. The plastic cellular states of liver cells: are EpCAM and Lgr5 fit for purpose? Hepatology 64, 652–662 (2016).
Michalopoulos, G. K. The liver is a peculiar organ when it comes to stem cells. Am. J. Pathol. 184, 1263–1267 (2014).
Forbes, S. J. & Rosenthal, N. Preparing the ground for tissue regeneration: from mechanism to therapy. Nat. Med. 20, 857–869 (2014).
Hall, C. et al. Regulators of cholangiocyte proliferation. Gene Expr. 17, 155–171 (2017).
Lazaridis, K. N. & LaRusso, N. F. The cholangiopathies. Mayo Clin. Proc. 90, 791–800 (2015).
Tanaka, E. M. & Reddien, P. W. The cellular basis for animal regeneration. Dev. Cell 21, 172–185 (2011).
Jin, S. G. et al. 5-Hydroxymethylcytosine is strongly depleted in human cancers but its levels do not correlate with IDH1 mutations. Cancer Res. 71, 7360–7365 (2011).
Thomson, J. P. et al. Loss of Tet1-associated 5-hydroxymethylcytosine is concomitant with aberrant promoter hypermethylation in liver cancer. Cancer Res. 76, 3097–3108 (2016).
Kafer, G. R. et al. 5-Hydroxymethylcytosine marks sites of DNA damage and promotes genome stability. Cell Rep. 14, 1283–1292 (2016).
Broutier, L. et al. Culture and establishment of self-renewing human and mouse adult liver and pancreas 3D organoids and their genetic manipulation. Nat. Protoc. 11, 1724–1743 (2016).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Krueger, F. & Andrews, S. R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572 (2011).
Wu, H., Caffo, B., Jaffee, H. A., Irizarry, R. A. & Feinberg, A. P. Redefining CpG islands using hidden Markov models. Biostatistics 11, 499–514 (2010).
Antonica, F., Orietti, L. C., Mort, R. L. & Zernicka-Goetz, M. Concerted cell divisions in embryonic visceral endoderm guide anterior visceral endoderm migration. Dev. Biol. 450, 132–140 (2019).
Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014).
Aloia, L. et al. Zrf1 is required to establish and maintain neural progenitor identity. Genes Dev. 28, 182–197 (2014).
McLean, C. Y. et al. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 (2010).
Stempor, P. & Ahringer, J. SeqPlots—interactive software for exploratory data analyses, pattern discovery and visualization in genomics. Wellcome Open Res. 1, 14 (2016).
Marshall, O. J. & Brand, A. H. damidseq_pipeline: an automated pipeline for processing DamID sequencing datasets. Bioinformatics 31, 3371–3373 (2015).
Kleinjan, D. A. et al. Long-range downstream enhancers are essential for Pax6 expression. Dev. Biol. 299, 563–581 (2006).
Acknowledgements
M.H. is a Wellcome Trust Sir Henry Dale Fellow and is jointly funded by the Wellcome Trust and the Royal Society (grant no. 104151/Z/14/Z). A.H.B. was funded by the Wellcome Trust Senior Investigator Award 103792 and a Royal Society Darwin Trust Research Professorship. S.J.F. is supported by a Medical Research Council (MRC) grant (grant no. MR/P016839/1). L.A. was supported by a Marie Skłodowska-Curie Postdoctoral fellowship (grant no.702585-EPILIPRO-H2020-MSCA-IF-2015) and a NC3Rs grant awarded to M.H. (grant no. NC/R001162/1). M.A.M. was supported by a MRC doctoral training grant (grant no. MR/K50127X/1). L.C.-E. was jointly funded by a Wellcome Trust Four-Year PhD Studentship with the Stem Cell Biology and Medicine Programme and a Wellcome Cambridge Trust Scholarship. J.v.d.A. was supported by a EMBO Long-term Fellowship (grant no. ALTF 1600_2014) and Wellcome Trust Postdoctoral Training Fellowship for Clinicians (grant no. 105839). F.A. was supported by an ERC advanced research grant to M.Z.-G. M.Z.-G. is a Wellcome Trust Senior Research Fellow. This work was partially funded by a H2020 LSMF4LIFE (grant no. ECH2020-668350) awarded to M.H., a ERC advanced grant to M.Z.G., a Wellcome Trust Senior Investigator Award awarded to E.A.M. (grant nos 104640/Z/14/Z and 092096/Z/10/Z) and a Cancer Research Programme Grant awarded to E.A.M. (grant nos C13474/A18583 and C6946/A14492). G.V. would like to thank Wolfson College at the University of Cambridge and the Genetics Society, London for financial help. The authors acknowledge core funding to the Gurdon Institute from the Wellcome Trust (grant no. 092096) and CRUK (grant no. C6946/A14492). The authors thank R. Krautz and W. Sanseverino for their advice on bioinformatic analyses, R. Arnes-Benito and A. A. Malcom for assistance with histological and immunostaining analyses, W. Reik and J. Silva for sharing TET1 plasmids, R. Butler for developing macro scripts, K. Harnish and C. Bradshaw of the Gurdon Institute’s genomic and bioinformatic facility for high-throughput sequencing, the Gurdon Institute facilities for assistance with imaging, animal care and bioinformatics analysis, A. Riddell and M. Paramor (Cambridge Stem Cell Institute) for assistance with FACS sorting and library preparation, respectively, the CRUK CI genomic facility for sequencing of the WGBS and RRHP libraries and M. Keighren (MRC Human Genetics Unit, University of Edinburgh) for technical support. M.H. would like to thank “Life Science Editors” for assistance during manuscript preparation and B. Simons and H. Clevers for their critical comments on the manuscript.
Author information
Authors and Affiliations
Contributions
M.H. and L.A. conceived and designed the project and interpreted the results. L.A., M.A.M., L.C.-E., G.B., G.V., N.A., J.v.d.A., A.R. and M.H. designed and performed experiments and interpreted results. L.A. designed and performed the in vitro experiments involving molecular biology techniques and organoid cultures, M.A.M. designed and performed the in vivo experiments and related stainings, L.C.-E. performed the hydroxymethylation and EdU stainings, G.B. performed the experiments with small molecule inhibitors. G.V. and E.A.M. prepared and analysed the WGBS and RRHP libraries, analysed the RNA sequencing and interpreted the corresponding bioinformatic analyses. N.A., A.R. and S.J.F. performed experiments with the β1-integrin model and interpreted the results of the p21 models. J.v.d.A. and A.H.B. performed the DamID-seq experiments. B.F.-C. helped with the in vivo analyses. R.A.C. helped with the bioinformatics analyses. R.L.M. provided the R26Fucci2a line. F.A. and M.Z.-G. performed the live imaging of ductal cells. L.A. and M.H. wrote the manuscript. All of the authors commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Non-proliferative EpCAM+ ductal cells initiate organoid cultures.
a, EpCAM+ ductal cells were isolated from WT livers by FACS using a sequential gating strategy as follows: cells were gated for FSC and SSC and subsequently singlets were gated using FSC/Pulse width. Then, cells were negatively selected for PE/Cy7 (to exclude CD11b+, CD31+ and CD45+ cells) and positively selected for APC (EpCAM+) to obtain CD11b−/CD31−/CD45−/EpCAM+ ductal cells (EpCAM+ cells). These cells give rise to proliferative organoids with ~15% efficiency. Representative bright field pictures of 500 EpCAM+ and EpCAM− cells 6 days after seeding. Graph represents mean ± SD of n = 3 independent experiments. b, RT-qPCR analysis of gene expression of the proliferation marker mKi67 (left) and stem-cell (Lgr5) and ductal (Epcam and Sox9) markers (right) at the indicated time points after seeding. Graphs represent the mean of n = 3 independent experiments. p-value obtained using Student’s two tailed t-test upon comparison to t = 0 h. *, p < 0.05; ***, p < 0.001. c, Proliferation analysis. EdU (10 μM) was incorporated to sorted EpCAM+ ductal cells at different intervals after seeding (0 h, 24 h and 48 h, arrows) and evaluated by immunofluorescence analysis 24 h after each incorporation. Representative images are shown. Scale bar, 10 μm. Graph represents the percentage of EdU + cells. Results are expressed as mean ± SD cells from n = 3 independent experiments. Student’s two tailed t-test statistical analyses were performed versus t = 24 h. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Extended Data Fig. 2 Transcriptional changes in ductal cells in vitro during liver organoid formation and in vivo upon damage.
a-e, RNA-seq analysis of ductal cells isolated from adult livers (0 h) and at different time points after culture. For DE genes, a pairwise approach with Wald test was performed on each gene using Sleuth. FDR < 0.1 was selected as threshold. a, Graphs represent the number of significantly DE genes for each comparison. b, Hierarchical clustering analysis of epigenetic regulators found DE (383 out of 698 published in ref. 37), in at least one comparison. Heatmap represents averaged TPM values scaled per gene. Results are presented as the averaged gene expression of the biological replicates. n, number of replicates. c-e, RNA-seq analysis of ductal cells isolated from adult livers (0 h) and at day 3 and day 5 after liver damage (2 mice were assessed per time point). c, The heatmap shows the 1552 DE genes at least in one comparison (TPM > 5, FDR < 0.1, |b| > 0.58). Clustering analysis identified 5 different clusters (Clusters 1-5) according to the expression profile (Cluster 1 n = 835; Cluster 2 n = 185; Cluster 3 n = 503; Cluster 4 n = 20; Cluster 5 n = 9). Number of genes in each cluster is indicated in brackets. Results are presented as average of the at least 3 biological replicates. d, Graph represents the number of significant DE genes in the different comparisons. e, GO and statistical analyses of the 3 main clusters identified in c were performed using DAVID 6.8.
Extended Data Fig. 3 TET1 catalytic activity is required for liver organoid formation and maintenance.
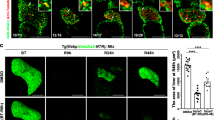
a, Tet1 and Lgr5 mRNA levels (n = 3 mice). Student’s two-tailed t-test statistical analyses were performed versus undamaged. b, Tet1 mRNA levels (24 h after transfection) and organoid formation efficiency 10 days after Tet1 siRNA knock-down using 4 independent Tet1 siRNAs. Data is presented as percentage relative to siCtrl. Graph indicates mean ± SD of n = 3 independent experiments. Student’s two-tailed t-test statistical analyses were performed versus mock-transfected cells. c, Scheme of the two different Tet1 alleles used. d, Tet1 mRNA levels in WT, Tet1hy/+ and Tet1hy/hy and Tet1 conditional knock-out (cKO) organoids presented as mean ± SD of n = 3 experiments. e, Representative Western blot image showing TET1 protein levels in WT, Tet1hy/+ and Tet1hy/hy organoids (3 independent experiments). f, Organoid formation efficiency from FACS-sorted EpCAM+ cells derived from RosaCreERT2 x Tet1 flx/flx livers treated with 5 μM hydroxytamoxifen (mean ± SD of n = 3 independent experiments). Student’s two-tailed t-test statistical analyses were performed versus non-induced control. g, Whole mount immunofluorescence staining of 5hmC (green) on WT, Tet1hy/hy, hypo-OE and hypo-OEcat.mut. organoids. Representative images are shown (2 experiments). Scale bar, 50 μm. h, Graph represents organoid size at the indicated passages (mean ± SD of n = 3 independent experiments). Student’s two tailed t-test statistical analyses were performed versus WT. i, Growth curves. j, Organoid formation efficiency at the indicated passage expressed as a percentage of organoids. Graphs represent mean ± SD of n = 3 independent experiments. Student’s two tailed t-test statistical analyses were performed versus WT. k, Representative confocal images of Cleaved Caspase 3 whole mount immunostaining on WT, Tet1hy/hy, hypo-OE and hypo-OEcat.mut. organoids (2 independent experiments). Scale bar, 25 μm.
Extended Data Fig. 4 WGBS of ductal cells uncovers a global epigenetic remodelling of the DNA methylome upon tissue damage.
a, Number of WGBS unique mapped reads in the different biological replicates. b, Bisulfite conversion rate. c-h, WGBS analyses were performed in merged biological replicates per time point (n = 2). Only CpG sites with ≥3 reads were further analysed. c, CpG counts in merged biological replicates per time point. d, Genome-wide Spearman’s correlation score at the time points analysed shows dynamic CpG modifications. e, Functional localization of DMRs. DMRs were called if the difference in cytosine modification between samples was ≥25% with a p-value of <0.05, using DSS software. f, Violin plot of the DMR length distribution (in base pairs) identified in the n = 2 biological replicates. Lines and numbers, median. g, Density plot indicating the difference in mCpG levels for loss/gain DMRs for each comparison. h, Venn diagram showing the overlap between TET1 targets (see Fig. 5) that are transcriptionally up-regulated and genes showing either loss (left) or gain (right) of mCpG at the TSS according to the WGBS analyses. Hierarchical clustering analyses of the overlapping genes are presented as heatmaps of TPMs scaled per gene (Z-score).
Extended Data Fig. 5 5hmC levels increase in ductal cells in vitro and in vivo upon damage.
a-c, EpCAM+ ductal cells sorted from DDC-treated WT livers (a), β1 integrin mutant mice fed with normal chow (undamaged) or DDC (b) or WT undamaged livers and grown in organoid culture conditions (c). 5hmC fluorescence intensity was normalized to DAPI. Data are presented as violin plots of the ratio 5hmC/DAPI. Each dot represents the median value of cells counted/mouse. Global median levels are shown in red. a, 353 cells from n = 4 undamaged mice, 231 cells from n = 5 mice after 3 days of DDC, and 392 cells from n = 5 mice at DDC d5; b, 138 cells from undamaged, 119 cells at day 1, 247 at day 7 and 125 at day 14 after returning the mice to normal chow (recovery) pooled from 2 livers isolated independently from 2 mice were analysed; c, 2500 (0 h), 900 (24 h) and 2000 (48 h) cells from n = 3 independent experiments were analysed. p-values were calculated using pairwise comparisons with Wilcoxon rank sum test. a, d3 versus d0 p = 1 × 10−13; d5 versus d0 p < 2.2 × 10−16. c, 0 h versus 24 h p < 2.2 × 10−16; 48 h versus 0 h p < 2.2 × 10−16. Scale bar, 10 μm. d, All 5hmC sites identified by RRHP. e, Number of genes associated to TSS showing differential 5hmC levels. The number of CpG sites (n) with unique gain of hydroxymethylation is shown. f, Graphs represent distribution of percentage of mCpG identified by WGBS in CGI outside TSS (n = 32673) using the average of the 2 independent samples (violin plots, black lines median, left) and number of 5hmC counts (median ± IQR) in CGI outside TSS (n = 25579) (right). g, GO and statistical analyses of the clusters identified in Fig. 4j (Cluster 2 n = 347; Cluster 3 n = 1659; Cluster 4 n = 1424; Cluster 6 n = 140) were performed using DAVID 6.8. Heatmap shows the expression profile of the 84 overlapping genes and is presented as averaged Z score of the 2 biological replicates.
Extended Data Fig. 6 TET1 regulates actively transcribed genes in liver organoids.
a-d, DamID-sequencing was performed in EpCAM+ sorted ductal cells derived from already established liver organoids (3 independent experiments). Only TET1-Dam peaks identified in all 3 experiments were considered for further analyses. a, Scheme of DamID-seq protocol. b, Heatmaps showing TET1 peaks identified by DamID-seq (left panels) and H3K4me3 peaks identified by ChIP-seq (right panels). Heatmaps are centred in the middle of the peak (0) and show a genomic window of ± 10 kb. Top heatmaps represent common peaks between TET1 and H3K4me3 (2848 peaks) while bottom heatmaps represent TET1-specific peaks (2254 peaks). c, Pie-chart indicates the percentage of genomic distribution of TET1-Dam peaks. d, GO and statistical analyses of biological processes among TET1-Dam targets in liver organoids were performed using DAVID 6.8. n, number of genes. e, 5hmC and 5mC levels determined by MeDIP and hMeDIP followed by qPCR on the indicated genomic region surrounding Lgr5 TSS in WT (black), Tet1hypo/hypo (blue) and hypo-OE (red) organoids. Graphs represent mean of n = 3 independent experiments. Student’s two tailed was performed comparing samples to WT. *, p < 0.05; ** = p < 0.01 f, TET1 ChIP-qPCR at Lgr5 TSS (left panel) and Lgr5 mRNA levels (right panel) in WT, Tet1hypo/hypo and hypo-OE organoids. Graphs represent mean ± SD of n = 3 independent experiments. Student’s two tailed t-test statistical analyses were performed versus WT. **, p < 0.01 g, Sorted EpCAM+ cells from WT livers were cultured in organoid culture conditions and harvested for DNA, chromatin and mRNA expression analyses at the indicated time points. Graphs represent mean of n = 3 independent experiments. Student’s two tailed t-test analyses were performed versus t = 0 h *, p < 0.05; ** = p < 0.01; *** = p < 0.001.
Extended Data Fig. 7 Treatment with Rapamycin impairs organoid formation.
a, EpCAM+ ductal cells freshly isolated from the undamaged liver were treated at 0-18hrs or 18-48hrs with the indicated small molecule inhibitors. Organoid formation was quantified at day 6. Graph represents organoid formation efficiency and indicates mean ± SD of n = 3 independent experiments. Statistical analyses were performed with two-ways ANOVA with Bonferroni’s multiple compared test (versus DMSO control group). DMSO control quantifications are shown in Fig. 6f. Representative pictures of organoids treated with the inhibitors at 18-48hrs are shown.
Extended Data Fig. 8 TET1 hypomorphic mice exhibit impaired ductal regeneration upon damage.
a, Graph represents mean ± SD of mouse weight of WT (n = 21 mice), Tet1hy/+ (n = 13 mice) and Tet1hy/hy (n = 27 mice) littermates. Student’s two tailed t-test statistical analyses were performed. b, Relative mouse weight of WT (n = 5), Tet1hy/+ (n = 1) and Tet1hy/hy (n = 5) mice. c, Representative H&E stainings (3 experiments) of intestines from 50 week old WT and Tet1hy/hy mice. Scale bar, 100μm. d, Representative H&E stainings (3 experiments) of small intestine from 10 week old WT and Tet1hy/hy mice treated with DDC for 5 days. Scale bar, 100μm. e-f, Box-and-whisker plots showing median and IQR of proliferating ductal cells (OPN+/Ki67+) during recovery (n = 3 WT and n = 4 Tet1hy/hy mice) (e) or total ductal cells (OPN+) at the different time points indicated (f) (Undamaged, n = 3 WT and n = 3 Tet1hy/hy mice; DDC, n = 7 WT and n = 6 Tet1hy/hy mice; Recovery, n = 3 WT and n = 4 Tet1hy/hy mice). Grey dots, outliers from a single counted FOV defined as > 1.5 IQR above or below the median. Red squares, median level corresponding to each independent mice. p-values obtained by two-sided Kolmogorov-Smirnov test. g, Population distribution of the total number of ductal cells (OPN+) Dashed lines show median values obtained from 55 FOV for WT (3 mice) and 56 FOV for Tet1hy/hy (3mice) at day 0 (undamaged) and 110 FOV for WT (3 mice) and 153 FOV for Tet1hy/hy (4 mice) at day 12 (recovery). h, PCK immunohistochemistry (3 experiments) from WT (left) and Tet1hy/hy (right) undamaged or in recovery after DDC (day 12) livers. Nucleus, Haematoxylin. Scale bar, 100 μm. i, Lgr5 and Tet1 mRNA levels, TET1 ChIP and hMedIP on Lgr5 TSS were analysed in undamaged and DDC treated livers. Graphs represent mean ± SD of values obtained from n = 3 independent biological replicates (dot). p-value was calculated using Student’s two-tailed t-test.
Extended Data Fig. 9 Ductal specific Tet1 conditional deletion impairs cholangiocyte-mediated liver regeneration.
a, Schematic of the Prom1CreERT2/RosalslZsGreen/Tet1flx/flx mouse model. b, Representative immunofluorescence analysis (OPN+ red, ZsGreen+, green) of Prom1∆Tet1/ZsGreen and Prom1Tet1WT/ZsGreen upon tamoxifen treatment and injection of AAV8-TBG p21 (2 mice per genotype). Nucleus, Hoechst. Scale bar, 50 μm c, Representative immunofluorescence analysis of livers from Prom1Tet1WT/ZsGreen mice injected with AAV8-TBG p21 not receiving tamoxifen treatment (2 mice per genotype). Scale bar, 100 μm. d, Tet1 expression in EpCAM+/ZsGreen+ ductal cells isolated by FACS from Prom1∆Tet1/ZsGreen (n = 4) or Prom1Tet/ZsGreen (n = 4) livers derived from mice treated for 3-cycles of DDC and collected 12 days after damage. Graph represents mean ± SD of Tet1 expression expressed as a fold change compared to Prom1Tet1WT/ZsGreen. Student’s two tailed t-test statistical analyses were performed. ***, p < 0.001. e, Representative pictures of P21 immunohistochemistry analyses. Scale bar, 200 μm. f, Weight curves of mice undergoing AAV8-TBG-p21 injection followed by DDC treatment (mean ± 95%CI). g, TET1 ChIP-qPCR analyses on target genes in ZsGreen+/EpCAM+ ductal cells isolated from Prom1Tet1WT/ZsGreen DDC-treated livers for 5 days. Cells isolated from 3 mice littermates were pooled used for each independent experiment (n = 2). ND, not detected. h, Graph represents mean ± SD of mRNA expression of Tet1 and selected target genes (fold change versus WT undamaged) in EpCAM+ ductal cells isolated from undamaged (n = 2 per genotype) or day 5 DDC-treated livers (n = 3 per genotype) derived from Prom1TET1WT/ZsGreen (grey) or Prom1∆Tet1/ZsGreen (blue) mice. Statistical analysis was performed using Student’s two-tailed t-test compared to the Prom1TET1WT/ZsGreen value at the corresponding time point.
Supplementary information
Supplementary Table 1
List of mouse models used.
Supplementary Video 1
Time lapse movie of EpCAM+ ductal cells FACS-sorted from undamaged R26Fucci2a mouse livers which were embedded in matrigel and grown in organoid culture conditions for 72 h.
Supplementary Dataset 1
RNA-sequencing data.
Supplementary Dataset 2
WGBS and RRHP data.
Supplementary Dataset 3
TET1-DamID sequencing and H3K4me3 ChIP-sequencing data.
Supplementary Dataset 4
List of DE genes in vivo and merge with TET1 targets, WGBS and RRHP.
Supplementary Dataset 5
List of antibodies, primers and siRNA sequence.
Source data
Source Data Fig. 1
Statistical Source Data Figure 1
Source Data Fig. 2
Statistical Source Data Figure 2
Source Data Fig. 3
Statistical Source Data Figure 3
Source Data Fig. 3
Unprocessed Blots Figure 3
Source Data Fig. 4
Statistical Source Data Figure 4
Source Data Fig. 5
Statistical Source Data Figure 5
Source Data Fig. 6
Statistical Source Data Figure 6
Source Data Fig. 7
Statistical Source Data Figure 7
Source Data Fig. 8
Statistical Source Data Figure 8
Source Data Extended Data Fig. 1
Statistical Source Data Extended Data Figure 1
Source Data Extended Data Fig. 2
Statistical Source Data Extended Data Figure 2
Source Data Extended Data Fig. 3
Statistical Source Data Extended Data Figure 3
Source Data Extended Data Fig. 3
Unprocessed Blots Exteded Data Figure 3
Source Data Extended Data Fig. 4
Statistical Source Data Extended Data Figure 4
Source Data Extended Data Fig. 5
Statistical Source Data Extended Data Figure 5
Source Data Extended Data Fig. 6
Statistical Source Data Extended Data Figure 6
Source Data Extended Data Fig. 7
Statistical Source Data Extended Data Figure 7
Source Data Extended Data Fig. 8
Statistical Source Data Extended Data Figure 8
Source Data Extended Data Fig. 9
Statistical Source Data Extended Data Figure 9
Rights and permissions
About this article
Cite this article
Aloia, L., McKie, M.A., Vernaz, G. et al. Epigenetic remodelling licences adult cholangiocytes for organoid formation and liver regeneration. Nat Cell Biol 21, 1321–1333 (2019). https://doi.org/10.1038/s41556-019-0402-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-019-0402-6
This article is cited by
-
Ten-eleven translocation-2-mediated macrophage activation promotes liver regeneration
Cell Communication and Signaling (2024)
-
TET (Ten-eleven translocation) family proteins: structure, biological functions and applications
Signal Transduction and Targeted Therapy (2023)
-
Control of stem cell renewal and fate by YAP and TAZ
Nature Reviews Molecular Cell Biology (2023)
-
Resolving the conflicts around Par2 opposing roles in regeneration by comparing immune-mediated and toxic-induced injuries
Inflammation and Regeneration (2022)
-
DNA methylation maintenance at the p53 locus initiates biliary-mediated liver regeneration
npj Regenerative Medicine (2022)