Abstract
The hallmark of Parkinson’s disease (PD) is the loss of dopaminergic (DA) neurons in the brain. However, little is known about why DA neurons are selectively vulnerable to PD. We previously completed a screen identifying genes associated with the progressive degeneration of DA neurons. Here we describe the role of a previously uncharacterized gene, CG42339, in the loss of DA neurons using Drosophila Melanogaster. CG42339 mutants display a progressive loss of DA neurons and locomotor dysfunction, along with an accumulation of advanced glycation end products (AGEs) in the brain. Based on this phenotype, we refer to CG42339 as vexed. We demonstrate that vexed is specifically required within cortex glia to maintain neuronal viability. Loss of vexed function results in excessive activation of the innate immune response in the brain, leading to loss of DA neurons. We show that activation of the innate immune response leads to increased nitric oxide signaling and accumulation of AGEs, which ultimately result in neurodegeneration. These results provide further insight into the relationship between the role of the immune response in the central nervous system and how this impacts neuronal viability.
Similar content being viewed by others
Introduction
Parkinson’s disease (PD) is the most prevalent movement disorder throughout the world and the second most prevalent neurodegenerative disease1. Clinical manifestation of PD includes a wide range of progressive locomotor dysfunctions such as tremors, bradykinesia, and impaired motor coordination2,3,4. This disease is primarily caused by the loss of dopaminergic (DA) neurons located within the substantia nigra5,6. There is currently no cure for PD, and the mechanism by which DA neurons are lost remains unknown. With an aging population, the prevalence rate of PD is expected to increase significantly over the next several decades.
Heritable forms of PD are linked to mutations in several genes. The first gene to be associated with PD is α-Synuclein, which acts through a toxic gain-of-function mechanism and is the major component of Lewy Body inclusions7,8. Loss of function mutations in Parkin have been associated with PD9. Parkin, an E3 ubiquitin ligase, is phosphorylated by PINK1 (PTEN-induced putative kinase 1) to promote mitochondrial quality control through selective mitophagy10. Additionally, long-term exposure to pesticides such as rotenone and paraquat that induce oxidative stress are also associated with higher incidence rates of PD11. However, identifying what specifically renders DA neurons vulnerable to PD remains unclear. Therefore, finding genes that are involved in the maintenance of DA neurons is crucial in order to discover potential therapeutic targets for PD.
Drosophila melanogaster has served as a valuable genetically tractable model system used to help uncover the cellular and molecular mechanisms underlying neurodegenerative diseases such as PD12. The Drosophila brain contains ~100 DA neurons organized into distinct clusters that can be easily quantified13. These DA neurons in the Drosophila brain are vulnerable to similar factors in PD patients, including expression of α-Synuclein7,14, mutations in genes such as PTEN-induced kinase 1 (PINK1) and Parkin15,16,17 and exposure to chemicals such as rotenone18,19. Moreover, this loss of DA neurons is linked to locomotor dysfunction which can be similarly recapitulated using Drosophila, further highlighting the utility of Drosophila models of PD20,21,22.
We previously completed a genome-wide screen identifying genes involved in the maintenance of DA neurons in the Drosophila brain23. From this screen, we identified the previously uncharacterized gene CG42339, which contained single nucleotide polymorphisms (SNPs) that were highly associated with a progressive loss of DA neurons and locomotor dysfunction with age. Although these phenotypes were validated using a mutant allele for CG42339, specific knockdown in DA neurons did not impair DA neuron viability or locomotor activity23, suggesting that CG42339 could exert neuroprotective effects in a non-cell autonomous fashion.
Here we describe our results, uncovering the mechanism by which CG42339 promotes the maintenance of DA neurons. First, we discovered a significant accumulation of advanced glycation end products (AGEs) in the brains of CG42339 mutants with age, which aligns with the prediction that CG42339 could be involved in scavenger receptor activity24. AGEs are formed when reducing sugars react with free amino groups from proteins. These AGEs will accumulate over time, resulting in tissue damage25,26. Previous evidence suggests proteins such as α-Synuclein and tau are glycated in patients with neurodegenerative diseases like Alzheimer’s Disease (AD) and PD27,28. Studies regarding PD specifically observed increases in AGE accumulation which was accompanied by increased α-Synuclein aggregation in Lewy bodies29,30,31.
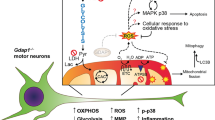
We next demonstrated that CG42339 is required in cortex glia, as tissue-specific knockdown results in the progressive loss of DA neurons. Since many types of glial cells are associated with the activation of the immune response, we also investigated the potential role of the innate immune response in the phenotypes found in CG42339 mutants. We found a significant increase in the activity of the innate immune response, as well as increased nitric oxide signaling in CG42339 mutant brains. It has been suggested that hyperactivation of innate immunity leads to neurodegeneration32,33. Brain inflammation in the central nervous system (CNS) can escalate the production of reactive oxygen species and reactive nitrogen species, which is linked to neurodegeneration, specifically PD34,35.
Finally, we demonstrated that both AGE accumulation and increased nitric oxide signaling are downstream of innate immune response activation. Altogether, these results further highlight the complex relationship between the immune response and the central nervous system and how these links can impact neurodegeneration.
Results
Neurodegeneration and locomotor dysfunction in CG42339 mutants
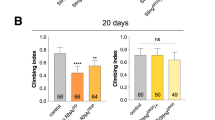
We previously identified CG42339 in a screen for genes associated with progressive degeneration of DA neurons23. To better understand the role of this previously uncharacterized gene, we expanded our analysis to all publicly available CG42339 mutant alleles. While homozygous mutants for each of these alleles displayed a normal number of DA neurons in the protocerebral posterior lateral-1 (PPL1) cluster on Day 3, they each showed a significant decrease in PPL1 neurons by Day 21 relative to heterozygous controls (Fig. 1A–Y). To confirm that this progressive loss of DA neurons is due to the loss of CG42339 function, we also measured neuron viability in mutant alleles crossed to a deficiency that spans the CG42339 region. We found a similar loss of DA neurons in each condition compared to homozygous mutants, suggesting that disruption of CG42339 function results in progressive degeneration of DA neurons (Fig. 1A–Y).
A–Y Progressive loss of PPL1 neurons stained with anti-tyrosine hydroxylase (green). Neuronal loss was assessed in CG42339MI01691 (A–F), CG42339MI09347(G–L), CG42339MI12354 (M–R), and CG42339MI13384 (S–X). Each allele was assessed in both heterozygous and homozygous conditions, as well as over the deficiency BSC540 that spans this region. Images were taken at 20x magnification with Z stack slice interval of 1.00 μm zoomed to 3.5x. Z Average climbing ability was measured for each allele on Day 3 and Day 21. Individual data points are shown with black dots. Error bars represent the SEM. ****p < 0.0001; *p < 0.05; n.s. not significant using Brown–Forsythe and Welch ANOVA tests with post hoc Games–Howell’s multiple comparisons. Scale bar in X is 20 μm for A–X.
While our initial measurements focused on PPL1 neurons, we also assessed the neuronal viability of other DA neuron clusters in the Drosophila brain. We also found a progressive, age-dependent loss of DA neurons located in the protocerebral posterior medial 1/2 (PPM1/2) cluster (Supplementary Fig. 1), demonstrating that the role of CG42339 in maintaining DA neurons is not limited to one subset of neurons. Interestingly, mutations in CG42339 did not result in the loss of DA neurons located within the Protocerebral Posterior Medial 3 (PPM3) cluster (Supplementary Fig. 2), highlighting that not all DA neurons are similarly vulnerable to specific mutations.
Since the loss of DA neurons is often tied to locomotor dysfunction12,21, we also assessed the climbing ability in CG42339 mutants. Heterozygous controls for each mutant allele displayed normal locomotor activity at both Day 3 and Day 21, while homozygous flies showed a progressive loss of motor activity by Day 21 (Fig. 1Z). Similar to our findings with DA neuron viability, flies bearing a mutation along with the corresponding deficiency also showed a loss of motor activity (Fig. 1Z). Together, these results demonstrate that CG42339 is required to maintain DA neuron viability and locomotor activity with age.
Accumulation of advanced glycation end products in CG42339 mutant brains
Although no previous experimental data has been published for these alleles, sequence analysis suggests that CG42339 may be associated with scavenger receptor activity24. Scavenger receptors target a wide range of molecules, including bacteria, cellular debris, and advanced glycation end products (AGEs), and these receptors are often closely linked with the innate immune response36.
AGEs are formed by the excessive glycation of proteins, lipids, and nucleic acids, and the accumulation of AGEs is associated with several age-related diseases. Long-lived proteins, in general, are susceptible to glycation, including several proteins associated with neurodegenerative diseases27. To assess the potential role of CG42339 as a scavenger receptor, we measured the accumulation of AGEs in aged wild-type brains in comparison to those of CG42339 mutants. While there was only a sparse accumulation of AGEs in wild-type brains at Day 21 (Fig. 2A–D), we noted a widespread accumulation of AGEs in CG42339 mutants (Fig. 2E, F). Since much of this staining was found in regions near clusters of DA neurons, we also labeled these neurons with a nuclear-localized Green Fluorescent Protein (UAS-Stinger)37, to determine their proximity to one another. We found a significant accumulation of AGEs in the vicinity of DA neurons within the PPL1 cluster, including large particles that appear to have aggregated in this region (Fig. 2H–J). Interestingly, we found no significant accumulation of AGEs in the region of the brain surrounding PPM3 in vexed mutants compared to controls (Supplementary Fig. 3). This could explain why PPM3 neurons are not lost in vexed mutant brains. Together, these results demonstrate that CG42339 is required to promote the turnover of AGEs and prevent their accumulation in the nervous system. Since this phenotype of accumulated AGEs in CG42339 mutants is similar to that shown for the human receptor for advanced glycation end products (RAGE)38,39, we refer to CG42339 as vexed (vex).
A–H AGE accumulation in flies at Day 21 with DA-neuron specific expression of a nuclear-localized GFP (UAS-Stinger) in a wildtype (A–D) or CG42339 mutant (E–H) background. Arrows designate small puncta of AGEs observed throughout the brain. Arrowheads designate large aggregates of AGEs found within close proximity to PPL1 neurons. Images were taken at 63x magnification with a Z stack slice interval of 0.88 μm zoomed to 3.5x. I Average number of AGE particles found within an area of 1500 μm2 in both wildtype and CG42339 mutant brains. J Average area occupied by AGE staining within an area of 1500 μm2 in both wildtype and CG42339 mutant brains. Individual data points are shown with black dots. Error bars represent the SEM. ****p < 0.0001; ***p < 0.001; using a Student’s t-test. Scale bar in H is 5 μm for A–H.
Vexed is required in glial cells to maintain DA neurons and locomotor function
To discover the tissue(s) in which vexed function is required to maintain DA neurons, we used the Gal4/UAS system to perform tissue-specific knockdown of vexed using RNAi40. We previously showed that the knockdown of vexed in DA neurons did not result in neuronal loss23, suggesting that Vexed could be working in a non-cell autonomous fashion to protect these neurons. We first used tubulin-gal441 to knock down vexed ubiquitously and found a significant loss of DA neurons in both the PPL1 and PPM1/2 neurons (Fig. 3D and S; Supplementary Fig. 4). Similarly, to our results with vexed mutants, there was no loss of PPM3 neurons (Supplementary Fig. 5). Since ubiquitous knockdown of vexed shares, a similar phenotype to mutants, we next sought to identify specific cell types where Vexed is required. We hypothesized that Vexed could provide non-cell autonomous protection to neurons by acting in glial cells. When vexed was knocked down in all glia using repo-gal442, we found DA neuron loss at Day 21 (Fig. 3F, S), demonstrating that vexed is required in glial cells.
A–S Clusters of PPL1 neurons stained with anti-tyrosine hydroxylase (green) upon tissue-specific knockdown of vexed. Comparison of controls (A, B) to ubiquitous knockdown (C, D), and knockdown in all glia (E, F), subperineural glia (G, H), ensheathing glia (I, J), astrocytic glia (K, L), perineural glia (M, N), and cortex glia (O–R). Images were taken at 20x magnification with a Z stack slice interval of 1.00 μm zoomed to 3.5x. T Average climbing ability measured for each condition at Day 3 and Day 21. U–X Anatomical organization of cortex glia (green) surrounding the DA neurons (red). Y–B′ Higher magnification of images panels U–X highlighting cortex glia surrounding DA neurons in the PPL1 cluster. Images were taken with Z stack slice intervals set at 1.00 μm. Individual data points are shown with black dots. Error bars represent the SEM. ****p < 0.0001 using Brown–Forsythe and Welch ANOVA tests with post hoc Games–Howell’s multiple comparisons. Scale bar in J is 20 μm for A–R. Scale bar in X is 40 μm. Scale bar in B′ is 20 μm.
Glial cells carry out a number of vital functions in the Drosophila nervous system, many of which are analogous to those of mammalian glial cells43,44,45,46. To determine the type(s) of glial cells in which Vexed is required, we next knocked down expression in individual glial cell subtypes using specific Gal4 drivers. DA neuron viability was assessed using RNAi in perineural glia (85G01-Gal4)47, subperineural glia (54C07-Gal4)47, ensheathing glia (56F03-Gal4)47, astrocytic glia (alarm-Gal4)48, and cortex glia (77A03-Gal4)47 and (54H02-Gal4)47 (Fig. 3G–S). There was no progressive degeneration of DA neurons in the PPL1 and PPM1/2 clusters when vexed was knocked down in perineural, subperineural, ensheathing, or astrocytic glia. However, we did find a progressive loss of DA neurons upon knockdown of vexed in cortex glia using two independent drivers (Fig. 3O–S and Supplementary Fig. 3), highlighting the need for Vexed function within these cells to maintain neuronal viability. Knocking down vexed using the ubiquitous, pan-glia, or cortex glia drivers also resulted in a progressive loss of locomotor function by Day 21 (Fig. 3T), again recapitulating the mutant phenotypes.
Cortex glia contains processes that surround neuronal cell bodies all throughout the protocerebrum in the Drosophila brain49, including the areas where the clusters of dopamine neurons are located (Fig. 3U–B′). Together, these results highlight the neuroprotective role of vexed in cortex glia.
Neurodegeneration in vexed mutants is caused by the activation of the innate immune response
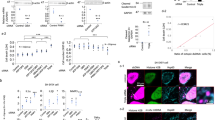
The relationship between the nervous system and the innate immune system has received growing attention recently, particularly in the context of neurodegenerative diseases. While the link between the innate immune response and neurodegeneration has been documented, it was only more recently determined that excessive activation of the innate immune response can itself result in neuronal loss50. As both glia activation and the accumulation of AGEs are linked to the innate immune response, we measured innate immune response activity in vexed mutant alleles as well as with RNAi knockdown in cortex glia. To assess activation of the innate immune response, we measured the expression levels of several antimicrobial peptides (AMPs) as a readout51,52,53. Vexed mutants displayed a significant increase in the levels of Attacin C, Drosomycin, Diptericin B, and Metchnikowin across all allelic variants at day 21 (Fig. 4A–D)54. In contrast, heterozygous vexed mutants that do not lose DA neurons showed no significant increase in AMP expression (Supplementary Fig. 6). Upon knockdown of vexed in the cortex glia, we tested the identical AMPs and found that there was an increase in expression for Drosomycin, Diptericin B, and Metchnikowin (Fig. 4E). These results demonstrate that the loss of vexed function results in a significant increase in innate immune activity in the nervous system. AMP expression is primarily determined by the activation of well-characterized transcription factors, including relish55,56. To determine whether the increase in AMP expression in vexed mutants is mediated by relish, we examined AMP levels in flies mutant for both vexed and relish. We found that double mutants did not show a significant increase in AMP expression at Day 21 (Fig. 4F), suggesting that the innate immune activity in vexed mutants requires relish activity.
A–F qPCR assessing transcript levels of several antimicrobial peptides from heads of vexed mutant alleles (A–D), RNAi-mediated knockdown of vexed in cortex glia (E), and flies mutant for both vexed and relish (F). G–O Measurement of PPL1 neuron viability in wildtype controls (G, H), vexed mutants (I, J), relish mutants (K, L), and flies with mutations in both vexed and relish (M, N). Images were taken at 20x magnification with a Z stack slice interval of 1.00 μm zoomed to 3.5x. Individual data points in each graph are shown with black dots. Error bars represent the SEM. ****p < 0.0001; ***p < 0.001; **p < 0.01; *p < 0.05; n.s. not significant using Brown–Forsythe and Welch ANOVA tests with post hoc Games–Howell’s multiple comparisons. Scale bar in N is 20 μm for G–N.
We next asked if the excessive innate immune response is responsible for the loss of DA neurons in vexed mutants. Consistent with our previous results, vexed mutants showed a progressive loss of PPL1 and PPM1/2 DA neurons by Day 21. However, relish mutants alone did not have a significant loss of neurons in these clusters. Interestingly, the loss of DA neurons from vexed mutations is prevented by introducing the relish mutation (Fig. 4G–O and Supplementary Fig. 7). Together, these results demonstrate that degeneration of DA neurons in vexed mutant brains is due to excessive activation of the innate immune response.
Enhanced nitric oxide signaling in vexed mutant brains
To better understand the mechanism responsible for the degeneration of DA neurons in vexed mutants, we next expanded our investigation to include signaling pathways that are associated with the accumulation of AGEs and innate immune activity. As nitric oxide activity is associated with these factors, we next measured nitric oxide activity in both wildtype and vexed mutant brains using DAR 4 M AM dye as a fluorescent nitric oxide indicator57. We found a significant increase in fluorescent signaling in aged vexed mutants compared to wild-type controls (Fig. 5A–C), showing that nitric oxide is enhanced in vexed mutants.
A–C Measurement of fluorescent intensity of DAR 4 M AM dye in wildtype (A) and vexed mutant (B) brains. D–L Viability of PPL1 neurons upon pharmacological inhibiting nitric oxide signaling with L-NAME. Conditions tested were wild-type controls raised on standard (D, E) or L-NAME-supplemented food (F, G) and vexed mutants raised on standard (H, I) or L-NAME-supplemented food (J, K). M–S Viability of PPL1 neurons upon genetically inhibiting nitric oxide signaling using a nos mutant allele. Conditions tested were wild-type controls (M, N), vexed mutants (O, P), and flies with mutations in both vexed and nos (Q, R). Images were taken at 20x magnification with a Z stack slice interval of 1.00 μm zoomed to 3.5x for (D–K) and (M–R). No further magnification (A, B). Individual data points in each graph are shown with black dots. Error bars represent the SEM. ****p < 0.0001; **p < 0.01; n.s. not significant using a Student t-test in C and a Brown–Forsythe and Welch ANOVA tests with post hoc Games–Howell’s multiple comparisons in L and S. Scale bar in B is 40 μm for A, B. Scale bar in K is 20 μm for D–K. Scale bar in R is 20 μm for M–R.
If enhanced nitric oxide signaling is associated with the loss of DA neurons in vexed mutants, we hypothesized that reducing nitric oxide signaling would be neuroprotective. To limit nitric oxide signaling pharmacologically, both wildtype and vexed mutants were raised on food supplemented with the nitric oxide inhibitor l-Name (l-nitro arginine methyl ester)58 or standard food. As shown previously, vexed mutants raised on standard food show a progressive loss of DA neurons (Fig. 5D–L and Supplementary Fig. 8). However, vexed mutants raised on l-NAME food maintain DA neurons in these clusters, demonstrating the neuroprotective effect of reducing nitric oxide signaling.
We also reduced nitric oxide signaling genetically using a mutation in nitric oxide synthase (nos) (NosMB04018)59. Similar to our results with l-NAME, we found that double mutants for vexed and nos maintained DA neurons at Day 21 (Fig. 5M–S). Together, these results demonstrate that the loss of DA neurons in vexed mutants is due to increased nitric oxide signaling, and that reducing nitric oxide signaling has neuroprotective effects.
Nitric oxide and AGE accumulation in vexed mutants depends on enhanced immune activity
We demonstrated that vexed mutants showing progressive loss of DA neurons and locomotor function also exhibit accumulation of AGEs, activation of the innate immune response, and nitric oxide signaling. To better understand these phenotypes and how they impact neurodegeneration, we next investigated any possible causal relationships between them.
To test the relationship between AGE accumulation and nitric oxide signaling, we measured AGE accumulation in the brains of both wildtype and vexed mutants that were raised on either standard media or media supplemented with l-NAME. If AGE accumulation depends on nitric oxide signaling, then we would expect the food supplemented with l-NAME to reduce AGE accumulation in vexed mutants. However, we found that raising vexed mutants on media supplemented with l-NAME had no impact on AGE accumulation, as the amount of AGEs found in vexed mutant brains was no difference between normal and l-NAME media (Fig. 6M–P, U, V). These results suggest that AGE accumulation is not a downstream consequence of nitric oxide signaling.
A–V Measurement of AGE accumulation in brains of wild-type flies raised on standard (A–D) or L-NAME-supplemented food (E–H), vexed mutant flies raised on standard (I–L) or L-NAME-supplemented food (M–P), and flies with mutations in both vexed and relish (Q–T). U Average number of AGE particles found within an area of 1500 μm2 in each condition. V Average area occupied by AGE staining within an area of 1500 μm2 in each condition. W–Y Fluorescent intensity of DAR 4 M AM dye in wild-type brains compared to flies with mutations in both vexed and relish. Z qPCR assessing transcript levels of antimicrobial peptides from heads of vexed mutants raised on L-NAME-supplemented food. Images were taken at 63x magnification with Z stack slice interval of 0.88 μm zoomed to 3.5x for (A–T). Images were taken at 20x magnification with a Z stack slice interval of 1.00 μm (W–X). Individual data points in each graph are shown with black dots. Error bars represent the SEM. ****p < 0.0001; ***p < 0.001, **p < 0.01; n.s. not significant using a Student t-test for Y and a Brown–Forsythe and Welch ANOVA tests with post hoc Games–Howell’s multiple comparisons for U, V, and Z. Scale bar in T is 4 μm for A–T. Scale bar in X is 40 μm for W, X.
We next assessed the relationship between AGE accumulation and activation of the innate immune response. If AGE accumulation depends on innate immune activation, then we would expect reduced AGE burden in the brains of flies mutant for both vexed and relish. We found that these double mutants showed a significant decrease in AGE burden compared to vexed mutants alone (Fig. 6Q–V), demonstrating that AGE accumulation in vexed mutants requires activation of the innate immune response.
The relationship between nitric oxide signaling and the innate immune response was assessed by measuring the intensity of DAR 4 M AM dye staining in the brains of flies mutant for both vexed and relish. If the enhanced nitric oxide signaling found in vexed mutants depends on the activation of the innate immune response, we would also expect these double mutants to have a similar dye intensity to controls. Indeed, we found that these double mutants no longer displayed an increase in nitric oxide signaling (Fig. 6W–Y), suggesting that nitric oxide signaling in vexed mutants also requires activation of the innate immune response.
Finally, we asked whether nitric oxide signaling is required for the activation of the innate immune response in vexed mutants. If enhanced nitric oxide signaling is required to activate the innate immune response, then we would expect vexed mutants raised on media supplemented with l-NAME to have reduced levels of AMP transcription. However, we found that AMP expression remained elevated when flies were raised on L-NAME food (Fig. 6Z). Altogether, these results demonstrate that activation of the innate immune response is required for both the accumulation of AGEs and enhanced nitric oxide signaling in vexed mutant brains.
Discussion
In this current study, we characterized the role of the previously unannotated gene vexed in the non-cell autonomous maintenance of DA neurons through the regulation of the innate immune response. We also found that DA neuron loss and locomotor dysfunction is accompanied by AGE accumulation and nitric oxide signaling in vexed mutants. Finally, we found that Inhibiting innate immunity led to DA neuron rescue, decreased AGE accumulation, and reduced nitric oxide signaling, suggesting that vexed mutations contribute to the hyperactivity of innate immunity, which stimulates AGE and nitric oxide accumulation.
The observation that Vexed exerts neuroprotective effects non-cell autonomously in cortex glia further highlights the importance of glia in the pathogenesis of neurodegenerative diseases. Recent investigations of glia in PD have garnered substantial attention. α-Synuclein is a protein that promotes synaptic vesicle trafficking, but when mutated, the protein will misfold, forming aggregates and bringing about the death of DA neurons60,61. Studies have suggested that the overactivation of glia promotes an increase in the aggregate formation of α-Synuclein62,63. Based on our findings and previous evidence, vexed may function in a similar manner or may enhance glia activation and impair other cellular processes.
Scavenger receptors perform a wide range of functions, including the removal of apoptotic cells and maintaining homeostasis by lipid transport. These receptors also clear the products of oxidative stress64,65. Our data and previous bioinformatic data suggest that Vexed likely acts as a scavenger receptor. Along with our results demonstrating that knockdown of Vexed in cortex glia is sufficient to induce activation of the innate immune response (Fig. 4), this suggests that defective scavenger activity of cortex glia induces the innate immune response. This idea is consistent with the observation that Advanced Glycation End Products (AGEs) are among common targets for scavenger receptors66 and the accumulation of AGEs we find in vexed mutant brains (Fig. 2).
Among the most well-studied scavenger receptors in Drosophila is Draper, which stimulates glial phagocytosis and clears apoptotic cells67,68,69,70. Evidence suggests that the upregulation of draper promotes the loss of neurons as well as locomotor dysfunction68. Specifically, draper was found to be expressed in cortex glia and cleared debris from dead neurons71. It will be interesting to examine in future studies whether Vexed may prevent AGE accumulation and oxidative stress normally through the cortex glia similarly to how Draper works.
Glial cells are the predominant phagocytes in the Drosophila central nervous system49. While this is most commonly associated with ensheathing glia48, cortex glia have been shown to phagocytose dead neurons in the developing optic lobe71.
The results from this study also highlight differences in AGE accumulation in various regions of the brain. While AGE accumulation is most prominent in regions surrounding DA neurons that are lost in vexed mutants, we did not find this accumulation in areas surrounding PPM3 neurons. Perhaps the anatomical location of these neurons renders them more resistant to this pathology. Although cortex glia clearly associates with DA neurons located within the PPL1 and PPM3 clusters (Fig. 3X), it is unclear whether cortex glia throughout the brain all react in an identical manner. Additionally, the use of fluorescent AMP reporters in previous studies did not show a uniform expression pattern in the central brain50. Differences in the induction of the innate immune response between regions of the brain could also explain differences in vulnerability.
Finally, it will also be interesting to examine the human ortholog of Vexed, Somatomedin-B, and thrombospondin type-1 domain-containing protein (SBS-PON). While most of the data regarding this gene comes from bioinformatic analysis72,73,74, it will also be of great interest to determine whether the neuroprotective effects found in our current study are conserved.
Methods
Fly stocks and husbandry
Drosophila melanogaster stocks were maintained at 25 °C on standard Drosophila media. Flies were collected at eclosion, separated by sex, and aged for either 3 days or 21 days at 29 °C. During the aging experiments, the flies were transferred to new media every 3 days. The following stocks were obtained from the Bloomington Drosophila Stock Center: CG42339MI01691 (#32773)75, CG42339MI13384 (#59652)75, CG42339MI12354 (#57935)75, CG42339MI09347 (#51278)75, UAS-CG42339IR (#67273)76, RelishE38 (#9458)77, Df BSC540 (#25068)78, y[1]w[1] (#1495)79, tubulin-Gal4 (#5138)41, repo-Gal4 (#7415)42, 85G01-Gal4 (#40436)47, 56F03-Gal4 (#39157)47, 54C07-Gal4 (#50472)47, alarm-Gal4 (#67032)48, 77A03-Gal4 (#39944)47, 54H02-Gal4 (#45784)47, UAS-Stinger (#84277)37, NosMB04018 (#24283)59.
Immunohistochemistry
Brains were dissected and stained as described previously23. Brains were dissected in 1X PBS and fixed for 20 min in 4% paraformaldehyde at room temperature. Brains were then washed four times using PBS with 0.3% Triton x-100 (0.3% PBST) for five minutes each. Samples were then placed in blocking buffer (PBS with 0.2% Triton x-100 and 0.1% normal goat serum) for a minimum of 1 h at 4 °C. After incubating with the blocking buffer, primary antibody was added and left on the sample for 48 h at 4 °C. Samples were then washed with 0.3% PBST four times for 5 min each at room temperature. Secondary antibodies were then added and samples were left to incubate at 2 h in the dark at room temperature. Samples were then washed with 0.3% PBST four times at 5 min each. Brains were then mounted using Vectashield mounting media (Vector Laboratories). Slides were imaged and then preserved at −20 °C. Primary antibodies used include rabbit anti-tyrosine hydroxylase (1:100, AB152, Millipore), rabbit anti-advanced glycation end products (1:100, ab23722, Abcam), rat anti-elav (1:20, Developmental Studies Hybridoma Bank), and chicken and-GFP (1:500, A10262, Thermo Fisher). Secondary antibodies used: Alexa Fluor 488 goat anti-rabbit, 568 goat anti-rabbit, 488 goat anti-rat (1:200, Fisher Scientific), and DAPI (1:1000).
Dopaminergic neuron quantification
DA neurons were counted using a Nikon Eclipse Ni-U fluorescent microscope equipped with a 20x objective. DA neurons of interest located in the protocerebral posterior lateral-1 (PPL1), Protocerebral posterior medial 1 and 2 (PPM1/2), and Protocerebral posterior medial 3 (PPM3) clusters were counted for both hemispheres within each individual brain sample. Brains were analyzed by genotype and sex for each experiment. A minimum of ten brains were analyzed for each group. Male and female brains were analyzed separately and then combined if there was no statistical difference between them. For mutants of CG42339 located on the X chromosome, only hemizygous males were analyzed. Experiments were performed in triplicate and were scored blindly with regard to genotype and condition.
Quantification of advanced glycation end products
AGE accumulation in brain samples was quantified using the Analyze Particles tool in FIJI (FIJI Is Just ImageJ)80. Z stacks of 48 slices (Interval of 0.88 μm) of the AGEs accumulating around the DA neurons were obtained and processed. The background was subtracted and the threshold was adjusted uniformly across all images. The total amount of particles and area were quantified over 6 images for each condition on both day 3 and day 21.
Image analysis
Microscopic images were taken using a Zeiss LSM 880 confocal microscope. A 20x objective was used to image neuronal clusters in each brain sample. AGE staining images were obtained using a 63x oil objective. All images, unless stated otherwise in the figure legend, are zoomed in to 3.5x. Confocal stacks were generated using parameters specifically outlined under each assay description in the figure legends. Brightness and contrast were then adjusted using FIJI (FIJI Is Just ImageJ)80 and Adobe Photoshop CC2020. All images were processed using the same parameters for brightness and contrast specific to each data set. Figures were then assembled in Adobe Illustrator CC2020.
Locomotor behavior
Flies were collected shortly after eclosion and separated by sex. Groups consisted of 10 male or female flies in each vial, with a total of 80–100 flies per genotype. Hemizygous males only were used for X chromosomal mutations. All other groups were combined since we saw no differences between males and females. We performed these experiments when the flies were aged to day 3 and day 21 at 29 °C. Flies were transferred onto new food every 3 days. We begin the climbing assay by transferring each group of flies into a tube that is made up of two glass vials connected at the open ends (total diameter, 2.5 cm; total height, 20 cm). Each group acclimated in the glass tube for 5 min. The climbing index is measured for each group by the percentage of flies that are able to climb to an 8 cm mark indicated on the glass vials in 20 s. The timer for these trials begins when the glass vial is tapped down onto a mouse pad. Three trials were carried out for each group of flies. After each trial, the flies were allowed 1 min to recover.
N-nitro-l-arginine methyl ester supplement
Standard fly media was prepared with 50 mM l-NAME supplemented as previously described58. Flies were collected after eclosion and placed on L-NAME-supplemented food or normal food as a control. Flies were maintained at 29 °C and were transferred to fresh food every 3 days. The sample size included a minimum of ten fly brains per each genotype.
Brain RNA isolation and quantitative PCR
RNA isolation and quantitative PCR was performed as previously described23. We isolated total RNA from 30–40 fly heads for each condition tested. Hemizygous males only were used for X chromosomal mutations. We used heterozygous females to evaluate transcript levels in Supplementary Fig. 6. The RNA extraction was carried out using Trizol (Invitrogen) followed by phenol-chloroform as per the manufacturer’s instructions. The total RNA for each sample was cleaned using the NEB RNA Cleanup kit (NEB T2030) according to the manufacturer’s instructions. We performed the qRT-PCR experiments using the Sybergreen Powerup master mix (Applied Biosystems) and the ABI7300 Real-Time Thermocycler. For each experiment, we performed three biological replicates per genotype and time point investigated. From this, we were able to obtain the average CT value for each biological replicate. Actin 5c was our internal control81. The fold change in expression relative to wildtype was found using the 2(−ΔΔCT) method for both mutants and RNAi knockdown experiments. The following primers were used as previously described:54
Actin 5c Forward primer 5′-CGAAGAAGTTGCTGCTCTGGTTGT-3′
Actin 5c Reverse primer 5′-GGACGTCCCACAATCGATGGGAAG-3′
Attacin C Forward primer 5′-CTGCACTGGACTACTCCCACATCA-3′, Attacin C Reverse primer 5′-CGATCCTGCGACTGCCAAAGATTG-3′, Cecropin A1 Forward primer 5′-CATTGGACAATCGGAAGCTGGGTG-3′, Cecropin A1 Reverse primer 5′-TAATCATCGTGGTCAACCTCGGGC-3′, Defensin Forward primer 5′-CCAGAGGATCATGTCCTGGTGCAT-3′ Defensin Reverse primer 5′-ACTTGGAGAGTAGGTCGCATGTGG-3′, Diptericin B Forward primer 5′-AGGATTCGATCTGAGCCTCAACGG-3′, Diptericin B Reverse primer 5′-TGAAGGTATACACTCCACCGGCTC-3′, Drosomycin Forward primer 5′-AGTACTTGTTCGCCCTCTTCGCTG-3′, Drosomycin Reverse primer 5′-CCTTGTATCTTCCGGACAGGCAGT-3′, Metchnikowin Forward primer 5′-CATCAATCAATTCCCGCCACCGAG-3′, Metchnikowin Reverse primer 5′-AAATGGGTCCCTGGTGACGATGAG-3′.
Nitric oxide signaling dye
The use of DAR 4 M AM nitric oxide signaling dye was adapted from ref. 57. Brains were dissected in 1X PBS. Each sample was then incubated in 10 μM DAR 4 M AM at room temperature (RT) in the dark for 1 h. Brains were then fixed for 20 min using 4% paraformaldehyde at RT. Samples were washed using 0.3% PBST four times at 5 min per wash. Brains were mounted using Vectashield media and immediately imaged. The sample size consisted of a minimum of 13 brains per genotype. Once imaged slides were stored at −20 °C.
Statistical analysis
Statistical analysis for the data were produced using Brown–Forsythe and Welch ANOVA tests with post hoc Games–Howell’s multiple comparisons or a Student’s t-test where appropriate using Graphpad Prism (Graphpad Software, Inc.).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The authors affirm that the conclusions of the article are present within the article, figures, and tables.
References
Beitz, J. M. Parkinson s disease a review. Front. Biosci. S6, 65–74 (2014).
Lesage, S. & Brice, A. Parkinson’s disease: from monogenic forms to genetic susceptibility factors. Hum. Mol. Genet. 18, R48–R59 (2009).
Postuma, R. B., Lang, A. E., Gagnon, J. F., Pelletier, A. & Montplaisir, J. Y. How does parkinsonism start? Prodromal parkinsonism motor changes in idiopathic REM sleep behaviour disorder. Brain 135, 1860–1870 (2012).
Poewe, W. Non-motor symptoms in Parkinson’s disease. Eur. J. Neurol. 15, 14–20 (2008).
Fearnley, J. M. & Lees, A. J. Ageing and Parkinson’s disease: substantia nigra regional selectivity. Brain 114, 2283–2301 (1991).
Vingerhoets, F. J. G. et al. Longitudinal fluorodopa positron emission tomographic studies of the evolution of idiopathic parkinsonism. Ann. Neurol. 36, 759–764 (1994).
Polymeropoulos, M. H. et al. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science 276, 2045–2047 (1997).
Spillantini, M. G. et al. Alpha-synuclein in Lewy bodies. Nature 388, 839–840 (1997).
Kitada, T. et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392, 605–608 (1998).
Matsuda, N. et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 189, 211–221 (2010).
Tanner, C. M. et al. Rotenone, paraquat, and Parkinson’s disease. Environ. Health Perspect. 119, 866–872 (2011).
Feany, M. B. & Bender, W. W. A Drosophila model of Parkinson’s disease. Nature 404, 394–398 (2000).
Mao, Z. Eight different types of dopaminergic neurons innervate the Drosophila mushroom body neuropil: anatomical and physiological heterogeneity. Front. Neural Circuits 3, 5 (2009).
Periquet, M., Fulga, T., Myllykangas, L., Schlossmacher, M. G. & Feany, M. B. Aggregated-synuclein mediates dopaminergic neurotoxicity in vivo. J. Neurosci. 27, 3338–3346 (2007).
Julienne, H., Buhl, E., Leslie, D. S. & Hodge, J. J. L. Drosophila PINK1 and parkin loss-of-function mutants display a range of non-motor Parkinson’s disease phenotypes. Neurobiol. Dis. 104, 15–23 (2017).
Greene, J. C. et al. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc. Natl Acad. Sci. USA 100, 4078–4083 (2003).
Yang, Y. et al. Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc. Natl Acad. Sci. USA 103, 10793–10798 (2006).
Radad, K., Rausch, W.-D. & Gille, G. Rotenone induces cell death in primary dopaminergic culture by increasing ROS production and inhibiting mitochondrial respiration. Neurochem. Int. 49, 379–386 (2006).
Coulom, H. Chronic exposure to rotenone models sporadic Parkinson’s disease in Drosophila melanogaster. J. Neurosci. 24, 10993–10998 (2004).
Cunningham, P. C., Waldeck, K., Ganetzky, B. & Babcock, D. T. Neurodegeneration and locomotor dysfunction in Drosophila scarlet mutants. J. Cell Sci. 131, jcs216697 (2018).
Whitworth, A. J. et al. Increased glutathione S-transferase activity rescues dopaminergic neuron loss in a Drosophila model of Parkinson’s disease. Proc. Natl Acad. Sci. USA 102, 8024–8029 (2005).
Tran, H. H. et al. Drosophila ubiquitin C-terminal hydrolase knockdown model of Parkinson’s disease. Sci. Rep. 8, 4468 (2018).
Davis, J. et al. Characterizing dopaminergic neuron vulnerability using genome-wide analysis. Genetics 218, iyab081 (2021).
Larkin, A. et al. FlyBase: updates to the Drosophila melanogaster knowledge base. Nucleic Acids Res. 49, D899–D907 (2021).
Kopytek, M., Ząbczyk, M., Mazur, P., Undas, A. & Natorska, J. Accumulation of advanced glycation end products (AGEs) is associated with the severity of aortic stenosis in patients with concomitant type 2 diabetes. Cardiovasc. Diabetol. 19, 92 (2020).
Oudes, A. J., Herr, C. M., Olsen, Y. & Fleming, J. E. Age-dependent accumulation of advanced glycation end-products in adult Drosophila melanogaster. Mech. Ageing Dev. 100, 221–229 (1998).
Li, J., Liu, D., Sun, L., Lu, Y. & Zhang, Z. Advanced glycation end products and neurodegenerative diseases: Mechanisms and perspective. J. Neurol. Sci. 317, 1–5 (2012).
Castellani, R., Smith, M. A., Richey, G. L. & Perry, G. Glycoxidation and oxidative stress in Parkinson disease and diffuse Lewy body disease. Brain Res. 737, 195–200 (1996).
Münch, G. et al. Crosslinking of α-synuclein by advanced glycation endproducts — an early pathophysiological step in Lewy body formation? J. Chem. Neuroanat. 20, 253–257 (2000).
Chen, L., Wei, Y., Wang, X. & He, R. Ribosylation rapidly induces α-synuclein to form highly cytotoxic molten globules of advanced glycation end products. PLoS ONE 5, e9052 (2010).
Dalfó, E. et al. Evidence of oxidative stress in the neocortex in incidental Lewy body disease. J. Neuropathol. Exp. Neurol. 64, 816–830 (2005).
Kounatidis, I. et al. NF-κB immunity in the brain determines fly lifespan in healthy aging and age-related neurodegeneration. Cell Rep. 19, 836–848 (2017).
Giunta, B. et al. Inflammaging as a prodrome to Alzheimer’s disease. J. Neuroinflammation 5, 51 (2008).
Jayaram, S. & Krishnamurthy, P. T. Role of microgliosis, oxidative stress and associated neuroinflammation in the pathogenesis of Parkinson’s disease: the therapeutic role of Nrf2 activators. Neurochem. Int. 145, 105014 (2021).
Mukherjee, S. Immune gene network of neurological diseases: Multiple sclerosis (MS), Alzheimer’s disease (AD), Parkinson’s disease (PD) and Huntington’s disease (HD). Heliyon 7, e08518 (2021).
Peiser, L., Mukhopadhyay, S. & Gordon, S. Scavenger receptors in innate immunity. Curr. Opin. Immunol. 14, 123–128 (2002).
Barolo, S., Carver, L. A. & Posakony, J. W. GFP and β-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. BioTechniques 29, 726–732 (2000).
Neeper, M. et al. Cloning and expression of a cell surface receptor for advanced glycosylation end products of proteins. J. Biol. Chem. 267, 14998–15004 (1992).
Bierhaus, A. et al. Diabetes-associated sustained activation of the transcription factor nuclear factor-κB. Diabetes 50, 2792–2808 (2001).
Brand, A. H. & Perrimon, N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415 (1993).
Lee, T. & Luo, L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22, 451–461 (1999).
Sepp, K. J., Schulte, J. & Auld, V. J. Peripheral glia direct axon guidance across the CNS/PNS transition zone. Dev. Biol. 238, 47–63 (2001).
Pereanu, W., Shy, D. & Hartenstein, V. Morphogenesis and proliferation of the larval brain glia in Drosophila. Dev. Biol. 283, 191–203 (2005).
Sonnenfeld, M. J. & Jacobs, J. R. Macrophages and glia participate in the removal of apoptotic neurons from theDrosophila embryonic nervous system. J. Comp. Neurol. 359, 644–652 (1995).
Auld, V. J., Fetter, R. D., Broadie, K. & Goodman, C. S. Gliotactin, a novel transmembrane protein on peripheral glia, is required to form the blood-nerve barrier in drosophila. Cell 81, 757–767 (1995).
Nickols, J. C., Valentine, W., Kanwal, S. & Carter, B. D. Activation of the transcription factor NF-κB in Schwann cells is required for peripheral myelin formation. Nat. Neurosci. 6, 161–167 (2003).
Jenett, A. et al. A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2, 991–1001 (2012).
Doherty, J., Logan, M. A., Tasdemir, O. E. & Freeman, M. R. Ensheathing glia function as phagocytes in the adult Drosophila brain. J. Neurosci. 29, 4768–4781 (2009).
Kremer, M. C., Jung, C., Batelli, S., Rubin, G. M. & Gaul, U. The glia of the adult Drosophila nervous system. Glia 65, 606–638 (2017).
Cao, Y., Chtarbanova, S., Petersen, A. J. & Ganetzky, B. Dnr1 mutations cause neurodegeneration in Drosophila by activating the innate immune response in the brain. Proc. Natl Acad. Sci. USA 110, E1752–1760 (2013).
Badinloo, M. et al. Overexpression of antimicrobial peptides contributes to aging through cytotoxic effects in Drosophila tissues. Arch. Insect Biochem. Physiol. 98, e21464 (2018).
Tzou, P. et al. Tissue-specific inducible expression of antimicrobial peptide genes in Drosophila surface epithelia. Immunity 13, 737–748 (2000).
Zhao, H. W., Zhou, D. & Haddad, G. G. Antimicrobial peptides increase tolerance to oxidant stress in Drosophila melanogaster. J. Biol. Chem. 286, 6211–6218 (2011).
Petersen, A. J., Rimkus, S. A. & Wassarman, D. A. ATM kinase inhibition in glial cells activates the innate immune response and causes neurodegeneration in Drosophila. Proc. Natl Acad. Sci. USA 109, E656–E664 (2012).
Petersen, A. J., Katzenberger, R. J. & Wassarman, D. A. The innate immune response transcription factor relish is necessary for neurodegeneration in a Drosophila model of ataxia-telangiectasia. Genetics 194, 133–142 (2013).
Ramesh, P., Dey, N. S., Kanwal, A., Mandal, S. & Mandal, L. Relish plays a dynamic role in the niche to modulate Drosophila blood progenitor homeostasis in development and infection. eLife 10, e67158 (2021).
Kozlov, A., Koch, R. & Nagoshi, E. Nitric oxide mediates neuro-glial interaction that shapes Drosophila circadian behavior. PLoS Genet. 16, e1008312 (2020).
Kraaijeveld, A. R., Elrayes, N. P., Schuppe, H. & Newland, P. L. l-Arginine enhances immunity to parasitoids in Drosophila melanogaster and increases NO production in lamellocytes. Dev. Comp. Immunol. 35, 857–864 (2011).
Bellen, H. J. et al. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167, 761–781 (2004).
Yasuda, T., Nakata, Y. & Mochizuki, H. α-Synuclein and neuronal cell death. Mol. Neurobiol. 47, 466–483 (2013).
Bendor, J. T., Logan, T. P. & Edwards, R. H. The function of α-synuclein. Neuron 79, 1044–1066 (2013).
Izco, M., Blesa, J., Verona, G., Cooper, J. M. & Alvarez-Erviti, L. Glial activation precedes alpha-synuclein pathology in a mouse model of Parkinson’s disease. Neurosci. Res. 170, 330–340 (2021).
Olsen, A. L. & Feany, M. B. Parkinson’s disease risk genes act in glia to control neuronal α-synuclein toxicity. Neurobiol. Dis. 159, 105482 (2021).
Patten, D. A. & Shetty, S. More than just a removal service: scavenger receptors in leukocyte trafficking. Front. Immunol. 9, 2904 (2018).
Canton, J., Neculai, D. & Grinstein, S. Scavenger receptors in homeostasis and immunity. Nat. Rev. Immunol. 13, 621–634 (2013).
Miyazaki, A., Nakayama, H. & Horiuchi, S. Scavenger receptors that recognize advanced glycation end products. Trends Cardiovasc. Med. 12, 258–262 (2002).
Awasaki, T. et al. Essential role of the apoptotic cell engulfment genes draper and ced-6 in programmed axon pruning during Drosophila metamorphosis. Neuron 50, 855–867 (2006).
Hakim-Mishnaevski, K., Flint-Brodsly, N., Shklyar, B., Levy-Adam, F. & Kurant, E. Glial phagocytic receptors promote neuronal loss in adult Drosophila brain. Cell Rep. 29, 1438–1448.e3 (2019).
Hilu-Dadia, R., Hakim-Mishnaevski, K., Levy-Adam, F. & Kurant, E. Draper-mediated JNK signaling is required for glial phagocytosis of apoptotic neurons during Drosophila metamorphosis. Glia 66, 1520–1532 (2018).
MacDonald, J. M. et al. The Drosophila cell corpse engulfment receptor draper mediates glial clearance of severed axons. Neuron 50, 869–881 (2006).
Nakano, R. et al. Cortex glia clear dead young neurons via Drpr/dCed-6/Shark and Crk/Mbc/dCed-12 signaling pathways in the developing Drosophila optic lobe. Dev. Biol. 453, 68–85 (2019).
Hasson, S. A. et al. High-content genome-wide RNAi screens identify regulators of parkin upstream of mitophagy. Nature 504, 291–295 (2013).
Fagerberg, L. et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteom. 13, 397–406 (2014).
Lopez, W. et al. Analysis of immune-related genes during Nora virus infection of Drosophila melanogaster using next generation sequencing. AIMS Microbiol. 4, 123–139 (2018).
Nagarkar-Jaiswal, S. et al. A library of MiMICs allows tagging of genes and reversible, spatial and temporal knockdown of proteins in Drosophila. eLife 4, e05338 (2015).
Perkins, L. A. et al. The transgenic RNAi project at Harvard Medical School: resources and validation. Genetics 201, 843–852 (2015).
Hedengren, M. et al. Relish, a central factor in the control of humoral but not cellular immunity in Drosophila. Mol. Cell 4, 827–837 (1999).
Cook, R. K. et al. The generation of chromosomal deletions to provide extensive coverage and subdivision of the Drosophila melanogaster genome. Genome Biol. 13, R21 (2012).
Biessmann, H. & Green, M. M. Molecular analysis of insertional mutations in the yellow gene region of Drosophila. J. Mol. Biol. 191, 573–576 (1986).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Dalui, S. & Bhattacharyya, A. Herbicide paraquat induces sex-specific variation of neuroinflammation and neurodegeneration in Drosophila melanogaster. Indian J. Biochem. Biophys. 51, 567–573 (2014).
Acknowledgements
The authors would like to thank members of the Babcock Lab for helpful discussions about the manuscript. This research was supported by the National Institutes of Health (R01NS110727 to D.T.B.).
Author information
Authors and Affiliations
Contributions
Designed research: J.D. and D.T.B. Performed research: J.D. and E.K. Analyzed data: J.D., E.K., and D.T.B. Wrote the paper: J.D. and D.T.B.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Davis, J., Kolaski, E. & Babcock, D.T. Vexed mutations promote degeneration of dopaminergic neurons through excessive activation of the innate immune response. npj Parkinsons Dis. 8, 147 (2022). https://doi.org/10.1038/s41531-022-00417-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41531-022-00417-5