Abstract
Cryo-electron microscopy (cryo-EM) has been widely used to reveal the structures of proteins at atomic resolution. One key challenge is that almost all proteins are predominantly adsorbed to the air-water interface during standard cryo-EM specimen preparation. The interaction of proteins with air-water interface will significantly impede the success of reconstruction and achievable resolution. Here, we highlight the critical role of impenetrable surfactant monolayers in passivating the air-water interface problems, and develop a robust effective method for high-resolution cryo-EM analysis, by using the superstructure GSAMs which comprises surfactant self-assembled monolayers (SAMs) and graphene membrane. The GSAMs works well in enriching the orientations and improving particle utilization ratio of multiple proteins, facilitating the 3.3-Å resolution reconstruction of a 100-kDa protein complex (ACE2-RBD), which shows strong preferential orientation using traditional specimen preparation protocol. Additionally, we demonstrate that GSAMs enables the successful determinations of small proteins (<100 kDa) at near-atomic resolution. This study expands the understanding of SAMs and provides a key to better control the interaction of protein with air-water interface.
Similar content being viewed by others
Introduction
Although cryo-EM has emerged as a powerful technique to reveal the high-resolution structures of biological macromolecules at close-to-native state, its current achievements still significantly fall short of their theoretical potential1. During the cryo-EM specimen preparation, the biomolecules are embedded in a thin liquid film and rapidly cooled down to liquid-nitrogen temperature. The interaction of biomolecules with air–water interface is widely recognized as a significant challenge, which limits successful reconstruction and achievable resolution1,2,3,4,5,6. The air–water interface often leads to the problems of preferential orientation and irreversible unfolding of protein particles. The unfolding of proteins at air–water interface is a thermodynamically downhill process and results in the denaturation of protein particles7.
To address this issue, a widely-used strategy is passivating the air–water interface using electron-transparent windows, like the surfactant molecules8,9,10,11,12 and affinity supporting films13,14,15,16,17,18,19,20,21,22. The use of surfactant has been long explored in preparing desirable cryo-specimen over the past three decades10,11. The amphiphilic surfactant can form the self-assembled monolayers (SAMs) at the air–water interface. These monolayers function as a protective barrier, preventing direct contact of the particles with the air–water interface and ensuring they remain in the liquid phase9. However, the choice of surfactant remains a try-and-error process, and very little is known about the interaction of protein with surfactant at the air–water interface. The protein particles can easily penetrate the fragile surfactant monolayers and eventually interact with the air–water interface1 (Fig. 1a). This may be a critical reason why optimization efforts for achieving the optimal surfactant yield varying results from one protein case to another, with none consistently performing well across various proteins. Besides, the presence of surfactant monolayers leads to contrast loss of images, posing a challenge in determining the structures of small biomolecules (<100 kDa), most of which are low-contrast and have not yet been resolved by cryo-EM3.
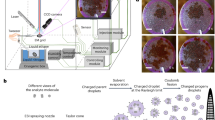
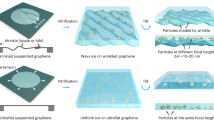
a Schematic illustration showing the surfactant self-assembled monolayers (SAMs) at the air–water interface are easily penetrated by the protein particles, leading to the preferential orientation of particles. b Particles with various orientations adsorbed on the impenetrable GSAMs superstructure, where surfactant SAMs lie flat on the graphene (Gr). The thickness of GSAMs (~0.9 nm) is thinner than that of surfactant monolayer (~2 nm) standing upright in (a). c, d Schematic illustrations showing the protein particles adsorbed on the GSAMs membrane with enriched orientations (d) compared to those on the pristine graphene surface (c).
Graphene grids have been adopted by many groups to address the air–water interface issue in cryo-EM. Various graphene functionalization strategies, such as low-energy plasma, ultraviolet/ozone treatment, and bioactive ligand modification16,17,18,19,20,21,22, have been developed to meet customized purposes during specimen preparation and have proven successful in various cases. However, the fabrication of high-quality and large-batch graphene grids remains a major barrier to their widespread application in the cryo-EM field.
In this work, we assemble the surfactants SAMs on the monolayer graphene membrane, to generate an impenetrable surfactant superstructure named GSAMs (Fig. 1b). Unlike conventional surfactant SAMs at the air–water interface, the surfactant molecules lie flat on the graphene surface (Fig. 1b), rendering the thickness of GSAMs membrane approximately 3 or 4 atoms and minimizing the background noise. The GSAMs membrane facilitates a more robust and straightforward graphene transfer process. In addition, the GSAMs membrane can efficiently anchor the proteins with enriched orientations at the GSAMs-water interface, alleviating the interface problems at both the air–water and graphene-water interfaces and improving the efficiency of cryo-EM analysis (Fig. 1c, d). This GSAMs superstructure enables us to achieve high-resolution reconstructions of several small biomolecules (<100 kDa), including a 2.6-Å resolution reconstruction of streptavidin (52 kDa), a 3.9-Å resolution reconstruction of human angiotensin-converting enzyme 2 (ACE2, 60 kDa), and a 3.3-Å resolution reconstruction of the complex formed by ACE2 and SARS-CoV-2 Spike protein receptor binding domain (ACE2-RBD, 100 kDa).
Results and discussion
Synthesis and characterization of free-standing GSAMs
To synthesize GSAMs, we first coated the surface of graphene/copper foil with stearic acid, one of the most common fatty acids in nature, which has been widely used in cleansers and soaps as a surfactant. This was simply achieved by dropping the solution (stearic acid dissolved in isopropanol) onto the graphene surface. After the evaporation of solvent, the SAMs of stearic acid were spontaneously formed (Fig. 2a). The molecular arrangements of SAMs on graphene were determined using scanning tunneling microscopy (STM) at a molecular resolution (Fig. 2b). The brighter stripes in the STM image represent the alkyl chains of stearic acid [CH3(CH2)16COOH], while the darker areas indicate the carboxyl groups. The assembled structure is correspondingly modeled in Fig. 2c, where the stearic acid molecules pack closely in a long-range order on the graphene surface. The periodic spacings of SAMs lattice are defined in two directions: one is ~19.2 Å, generated by the head-to-head interaction of adjacent molecules through the formation of two hydrogen bonds, and the other is ~4.2 Å, determined by calculating the average space of neighboring alkyl chains.
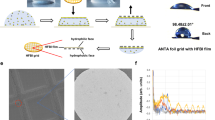
a Schematic illustration showing the on-site assembly of GSAMs superstructure. After the evaporation of surfactant solution (left), the self-assembled monolayers of surfactant molecules are spontaneously formed on the graphene/copper surface (middle). The free-standing GSAMs superstructure on the water surface can be achieved after the copper foil is etched away (right). b Molecular-resolution STM image of surfactant (C17H35COOH) monolayer, where the stripe features correspond to the alkyl chains of C17H35COOH. c The assembled structure model of C17H35COOH on the graphene surface, based on the parameters in (b). d Inch-size free-standing GSAMs membranes at the air–water interface. e Coverage statistics of suspended GSAMs on the TEM grid, the averaged coverage is ~99.5%. Inset: a representative SEM image of suspended GSAMs membranes. f Cryo-EM image of suspended GSAMs on the grid hole. The stripe-like patterns were clearly observed. g Fast Fourier Transform (FFT) of GSAMs. The diffraction patterns of SAMs superstructure indicated the stripe-like character with well-defined periodicities of 19.2 Å and 4.26 Å. h DFT simulations of stearic acid molecules arranged along the zigzag direction of graphene lattice. The chain spacing of C17H35COOH is 4.26 Å, in agreement with the experimental results. i Calculated electron diffraction patterns from (h).
Benefiting from this well-ordered structure, the inch-size GSAMs was robust and remained free-standing at the air–water interface after the underlying copper substrate was etched off (Fig. 2d). In contrast, the graphene without SAMs coating was cracked and folded into graphene scrolls (Supplementary Fig. 1). The free-standing GSAMs membrane facilitates the graphene transfer process. Compared with the conventional polymer-assisted transfer method (such as using polymethyl methacrylate), which is often an arduous process resulting in low-coverage and contaminated graphene surface23,24,25, the SAMs enable simpler and more practical graphene transfer onto the grids with a high yield (Supplementary Fig. 2, Supplementary Movie 1, “Methods”). The large-area suspended GSAMs membranes on the grid exhibit a near-perfect coverage of ~99.5% (Fig. 2e). The thickness and surface roughness of GSAMs were measured by atomic force microscopy (AFM). The thickness of GSAMs is ~0.9 nm (Supplementary Fig. 3a). Considering that graphene is atomically thin with a thickness of ~0.34 nm, the thickness of the SAMs layer is 0.5–0.6 nm, which indicates the presence of a monolayer of SAMs on the graphene surface. Furthermore, the suspended GSAMs membranes are flat, with an average surface roughness of ~0.8 nm (Supplementary Fig. 3b).
We then used cryo-EM to characterize the superstructure of suspended GSAMs. The well-aligned stripe-like patterns were clearly resolved at cryogenic temperature (Fig. 2f), which was absent in the traditional room-temperature EM characterization due to the irradiation damage (Supplementary Figs. 4 and 5). The corresponding Fast Fourier Transform (FFT) image displayed the diffraction patterns of graphene and additional twofold-symmetry diffraction spots associated with the SAMs (Fig. 2g). The twofold-symmetry spots indicated the molecules of SAMs were packed in a stripe-like order on the graphene, with the lattice parameters of 4.26 Å and 19.2 Å. These results were in good agreement with the STM characterizations (Fig. 2b, c).
We further performed theoretical simulations to investigate the interactions between SAMs and graphene. After structural optimization, the SAMs of stearic acid molecules formed and arranged as strip-like structures on graphene (Fig. 2h). The binding energy between graphene and SAMs was −199.02 mJ/m2, demonstrating the assembly process was spontaneous in thermodynamics. The alkyl chains of stearic acid were along the direction of graphene lattice, and the spacing of adjacent chain was 4.26 Å, the double of graphene lattice (2.13 Å) (Fig. 2h). Accordingly, the calculated twofold-symmetry diffraction spots of SAMs appeared with the same direction as graphene lattice (Fig. 2i). These theoretical simulations were in excellent agreement with our cryo-EM observations.
GSAMs membrane alleviates the air–water interface problem
GSAMs membranes, consisting of self-assembled amphiphilic surfactant molecules, have periodic functional groups of carboxyl and alkyl chains, offering diverse interaction manners with target protein particles. This well-designed structure may not only prevent the protein adsorption at the air–water interface but also provide a solution to the preferential orientation problem. To test this idea, we characterized the spatial distributions of 20S proteasome particles in the vitreous ice layer using cryo-electron tomography (cryo-ET). As shown in Fig. 3a, b, most proteasome particles adsorbed at the air–water interfaces when the cryo-specimen was prepared with traditional holey carbon support. We also noticed the presence of preferential orientation and protein debris at the air–water interfaces (Fig. 3c). When the protein particles were supported by GSAMs membrane, they tended to adsorb at the GSAMs-water interface (Fig. 3d, e), and showed diverse views of 20S proteasomes, including the side view (rectangle-shaped particle) and top view (ring-shaped particle) (Fig. 3f). In addition, the proportion of smaller particles, likely protein debris, was significantly lower at the GSAMs-water interface than at the air–water interface. These results revealed that the GSAMs could effectively avoid the air–water interface interference, and enrich orientations of proteins during the cryo-specimen preparation.
a–c Spatial distribution of protein particles on the holey carbon grid, which was commonly used in traditional cryo-specimen preparation protocol. a Schematic illustration showing most particles adsorb at the air–water interface. b Cryo-electron tomography (cryo-ET) reconstruction reveals the locations of particles in vitreous ice layer. Each red spot represents one individual 20S proteasome particle. c Typical slices extracted from (b) showing the particle adsorption at the air–water interfaces with preferential orientation and protein denaturation (marked in red cycles). d–f Spatial distribution of protein particles on GSAMs. The 20S proteasome particles on the GSAMs superstructure exhibit diverse orientations, including the side view and top view. g Typical cryo-EM images and representative 2D class averages (inset) showing 0.6 mg/mL 20S proteasome particles on the GSAMs with various orientations. h 2.7 mg/mL 20S proteasome in the presence of 8 mM CHAPSO exhibits denatured structures and low-density distributions. i 2.7 mg/mL 20S proteasome in the presence of 0.17 mM DDM shows limited orientations. The surfactants were slightly above their critical micelle concentrations. Scale bars, 50 nm.
We further compared the efficiency of GSAMs with other commonly-used surfactants in alleviating the air–water interface effect. We collected the single-particle cryo-EM dataset of 20S proteasome on the GSAMs, whose concentration was 0.6 mg/mL before being applied onto GSAMs. The protein particles exhibited high density on the GSAMs, displaying various orientations, including side view, top view, and tilted views in between (Fig. 3g). Moreover, we found multiple rounds of sample application and blotting with GSAMs grids26 could further reduce the concentration requirement (Supplementary Fig. 6). In preparing the cryo-EM specimens with surfactants, such as CHAPSO (3-(3-cholamidopropyl-dimethylammonio)-2-hydroxy-1-propanesulfonate) or DDM (dodecyl-β-D-maltoside), we observed that a high concentration of 2.7 mg/mL was necessary to achieve a suitable particle concentration in the vitreous ice (Fig. 3h, i). The addition of CHAPSO resulted in the clear observation of protein debris in cryo-EM micrographs, indicating potential interaction and damage by the air–water interface (Fig. 3h). Moreover, these particles showed a much lower density distribution compared to those on GSAMs (Fig. 3g). Limited orientation issue was also observed in the presence of DDM (Fig. 3i). We further tested the effects of GSAMs, CHAPSO and DDM on another protein, the spike protein (Supplementary Fig. 7). At a low concentration (0.6 mg/mL), we observed evenly distributed and high-density spike particles on GSAMs, whereas very few particles were seen in the presence of CHAPSO or DDM, even at a higher concentration (2 mg/mL). In addition, many small dots, likely protein debris, were observed with CHAPSO. These data suggest that the surfactants used as additives during cryo-EM specimen preparation do not function consistently well, and the protein particles could penetrate the protective surfactant monolayer at the air–water interface. Thus, we demonstrated that the impenetrable GSAMs membrane is much more efficient and robust in passivating the air–water interface.
GSAMs enriches the orientations of proteins
To better assess the role of surfactant SAMs in enriching protein orientations and exclude the effect of the underlying graphene, we compared the orientational distributions of various proteins on the GSAMs with those on pristine graphene, respectively. For the streptavidin protein with D2 symmetry, only side view of streptavidin particles appeared on the pristine graphene surface, while both top view and side view orientations emerged on the GSAMs (Supplementary Fig. 8). Similar results were obtained for the 20S proteasome with D7 symmetry. Particles on the GSAMs exhibited various orientations, including side view and top view (Fig. 4a), but showed preferential orientation of side view on the pristine graphene that was consistent with previous reports27 (Fig. 4b, Supplementary Fig. 9). We also calculated the orientational distribution efficiencies of the 20S proteasome with the addition of CHAPSO (Fig. 4c) and DDM (Fig. 4d), resulting in efficiencies of 0.82 and 0.12, respectively. Interestingly, the orientational distribution efficiency of the 20S proteasome with CHAPSO was higher than that on GSAMs, which was 0.52. This suggests that although CHAPSO did not protect some particles from air–water interface damage, the remaining particles adsorbed at the surfactant adopted a richer variety of orientations, consistent with other studies12.
a–d The Euler angle distributions and preferred views of 20S proteasome with the presence of GSAMs (a), graphene (b), surfactant CHAPSO (c) and surfactant DDM (d), respectively. e, g, i The Euler angle distributions and preferred views of Spike protein of SARS-CoV-2 (e), ACE2-RBD (g), and ACE2 (i) particles on the GSAMs, respectively. f, h, j The Euler angle distributions and preferred views of Spike protein of SARS-CoV-2 (f), ACE2-RBD (h), and ACE2 (j) particles on the pristine graphene, respectively. k, l The orientational distribution efficiencies (k) and particle utilization ratios (l) of 20S proteasome, Spike protein of SARS-CoV-2, ACE2-RBD, and ACE2 on the GSAMs (red) and pristine graphene (blue), respectively. The efficiency values for measurements of the 20S proteasome in the presence of CHAPSO (in purple) or DDM (in green) are also included in (k).
We further tested the orientational distributions of other macromolecules without symmetry on the GSAMs. During the COVID-19 pandemic, SARS-CoV-2 virus infects human beings through a high-affinity interaction between its Spike protein with ACE2 protein on mammalian cell membrane. We applied the Spike protein of SARS-CoV-2, ACE2 protein, and the complex of ACE2-RBD onto the GSAMs, and performed the single-particle cryo-EM analysis (Supplementary Table 1). Our findings revealed diverse orientational distributions for all these samples on the GSAMs (Fig. 4e, g, i). In contrast, the Spike protein exhibited a preferential orientation on the pristine graphene, and the orientational distributions of the other two proteins also differed from those observed on GSAMs (Fig. 4f, h, j, Supplementary Fig. 10). More specifically, we calculated the orientational distribution efficiencies of these proteins28. We discovered that there was a more even and efficient distribution of orientations for all these proteins when they were placed on GSAMs compared to pristine graphene (Fig. 4k).
To examine the capability of GSAMs in preserving the proteins’ native structures, we sought to determine the particle utilization ratio, which refers to the proportion of the final number of particles used for structural reconstruction relative to the total particle number initially obtained from the raw micrographs (Fig. 4l). We found that, for 20S proteasome, Spike protein, ACE2, and ACE2-RBD complex, the GSAMs generally yielded higher particle utilization ratio than the pristine graphene (Fig. 4l). Especially for the Spike protein and ACE2-RBD complex, which consist of multiple polypeptides, the particle utilization ratio on GSAMs (~0.722 for Spike and ~0.561 for ACE2-RBD) were significantly higher than those on pristine graphene membrane (~0.442 for Spike and ~0.187 for ACE2-RBD). These results suggest that the impenetrable surfactant monolayers on the GSAMs enhance cryo-EM structural reconstruction efficiency, likely due to the GSAMs alleviating the interface issues and enriching the protein orientations.
GSAMs enables the high-resolution cryo-EM reconstruction
The GSAMs superstructure, owing to its atomically thin thickness, is supposed to generate minimal background noise. Therefore, the GSAMs is particularly beneficial for cryo-EM reconstruction of macromolecules with small molecular weight. In our practice, we applied the GSAMs to the cryo-EM reconstruction of various macromolecules with molecular weight ranging from 700 kDa to 52 kDa.
The attempt to reconstruct the ACE2-RBD structure using a commonly-used holey carbon film failed due to limited orientations at the air–water interface (Fig. 5a). In contrast, successful reconstruction at a resolution of 3.3 Å was achieved on GSAMs with various orientations (Fig. 5b, c). Using GSAMs, we reconstructed the cryo-EM structure of 20S proteasome at a 2.0-Å resolution, enabling clear resolution of the hole in the benzene ring at such high resolution (Fig. 5c, Supplementary Fig. 11a, b). We were also able to successfully determine the cryo-EM structures of small macromolecules, such as ACE2 (60 kDa) and streptavidin (52 kDa), at resolutions of 3.9 Å and 2.6 Å, respectively (Fig. 5d, e, Supplementary Figs. 11c–f and 12). These results highlight the effectiveness of the GSAMs as a good supporting substrate for determining high-resolution cryo-EM structures of biomolecules, especially for those with small molecular weight (<60 kDa).
a, b Representative 2D class averages of ACE2-RBD on the conventional holey carbon film with preferential orientation (a) and GSAMs with various orientations (b), respectively. Scale bar in the 2D class averages, 5 nm. c Cryo-EM density maps of streptavidin (at a 2.5-Å resolution, 52 kDa), ACE2 (3.9 Å, 60 kDa), ACE2-RBD (3.3 Å, 100 kDa), Spike protein of SARS-CoV-2 (3.1 Å, 440 kDa) and 20S proteasome (2.0 Å, 690 kDa), reconstructed by using particles on GSAMs. The maps were colored by local-resolution distribution. d, e Selected densities from the two small proteins (<100 kDa), ACE2 map (d) and streptavidin map (e), with the corresponding atomic models docked.
In summary, we have successfully synthesized the inch-size GSAMs superstructure by assembling the surfactant monolayers on graphene membrane. We showcase the potential of impenetrable surfactant monolayers of GSAMs to alleviate the air–water interface problem in high-resolution cryo-EM analysis. The GSAMs membranes effectively accommodate various proteins, providing a key to getting better control of the interactions between protein and air–water interface. The GSAMs exhibits the ability to enrich the orientational distributions of various proteins, enabling high-resolution structural determination of macromolecules with wide-range molecular weights from 700 to 52 kDa. Thus, our work opens an avenue in cryo-EM analysis of biological samples in aspects of both fundamental understandings and technological applications. Beyond cryo-EM, we believe the GSAMs membranes are promising in the fields of drug delivery, separating membranes, and self-assembly science.
Methods
Preparation of high-quality graphene films
Graphene films with large single-crystal domains were prepared via chemical vapor deposition (CVD) method. 27-μm-thick Cu foils (Kunshan Luzhifa Electron Technology Co., Ltd., China) were placed in a homemade low-pressure CVD system, which is equipped with a 6-inch quartz tube and three heating zones. An asynchronous heating process and a temperature gradient were applied to the Cu foils, which promoted the enlargement of Cu single crystals29. Then the samples were annealed for 30 min in a mixture of Ar and O2 (PAr = 1000 Pa, PO2 = 0.4 Pa) and 10 min in a mixture of Ar and H2 (PAr = 500 Pa, PH2 = 500 Pa), respectively. The graphene growth proceeded under a gas mixture of H2 (PH2 = 500 Pa), Ar (PAr = 500 Pa), and CH4 (PCH4 = 1.6 Pa) for 90 min. The CVD chamber was cooled down to room temperature after growth procedure, during which the same atmosphere and partial pressures were maintained. The high-quality graphene films are also commercially available in Beijing Graphene Institute (BGI) company (http://www.bgi-graphene.com/type/100).
Preparation of free-standing GSAMs superstructure
Plasma treatment of Cu foil
To remove graphene on the back side of Cu foil, we put Cu foil into a reactive ion etcher (Pico SLS, Diener) with an airflow of 10 sccm. Then the air plasma was generated at a controlled power of 150 W. After a 3-min treatment, the backside graphene was fully removed.
Self-assembly of stearic acid monolayers
Stearic acid was dissolved and diluted in isopropanol (IPA) to reach the mass fraction of 0.005%. Then we used a capillary to drip the solution on graphene to fully cover its surface. Normally, each drop volume of stearic acid/IPA solution we use was about 10 μL, which could spread over an area of about 1.5 cm × 1.5 cm. After the evaporation of IPA, stearic acid monolayer was self-assembled on graphene surface.
Etching
The Cu/graphene/SAM composite was placed on the surface of 0.5 M (NH4)2S2O8 aqueous solution to etch the Cu foil away. After etching, the free-standing graphene/SAM superstructure was floated at the air–water interface.
Preparation of suspended GSAMs membranes
TEM grids (e.g., Quantifoil-R1.2/1.3, 300 mesh) were carefully positioned on the filter paper, which had been submerged into the etching solution in advance. The solution was then gradually pumped away to lower down the solution surface, making the floating GSAMs superstructure deposit onto TEM grids.
Rinsing
GSAM-coated TEM grids were transferred away from etching solution and submerged into deionized water to wash away the inorganic contaminations. The GSAMs grid could be rinsed in the isopropanol solution for 30 s to remove the organic contaminations if needed.
Drying
The suspended GSAMs membranes on the grids were air-dried in the cleanroom to avoid extra contaminants after being taken out from the deionized water.
Preparation of pristine graphene grids
The pristine graphene grids are prepared by a polymer-free clean transfer method22,30. Firstly, the commercial holey carbon grids (Quantifoil, Au 300 mesh R1.2/1.3) are placed on the surface of graphene/copper substrate. Then, an isopropanol solution is dropped onto these grids. After the isopropanol evaporates, the holey carbon grids will be tightly attached to the surface of the graphene/copper substrate. Subsequently, 1 M (NH4)2S2O8 aqueous solution is used to etch the copper substrate away, resulting in the transfer of graphene film onto the surface of holey carbon grids. After rinsed in deionized water and isopropanol solution, the clean graphene grids can be obtained.
Coverage of GSAMs grid
The GSAMs grid coverage is determined by calculating the ratio of the number of holes with intact membranes to the total number of holes in each grid square. We collected the SEM images of 104 grid squares from the GSAMs grid (Supplementary Fig. 2b), and each grid square has ~430 holes (Supplementary Fig. 2c). By counting the number of holes covered with broken, suspended GSAMs membranes, the GSAMs coverage can be obtained. The coverage statistics based on these 104 grid squares have been shown in the Fig. 2e.
Characterization of GSAMs by AFM
The surface roughness of suspended SAMs membranes was characterized by atomic force microscopy (AFM, Bruker Dimension Icon) in tapping mode with a commercial AFM tip (OTESPA-R3, Bruker company). The material, tip radius, spring constant, and resonance frequency of OTESPA-R3 AFM tips are 0.01–0.02 ohm-cm Silicon, ~7 nm, ~26 N/m, and ~300 kHz, respectively. The rate of data collection is 1 line/s for 256 lines. The values of surface roughness can be directly obtained from the AFM image.
Density functional theory (DFT) calculations
All the DFT calculations were performed using the Vienna Ab initio Simulation Package (VASP)31,32. The electron-ion and electron-electron interactions were described by the projector augmented wave (PAW) methods and Perdew-Burke-Ernzerh of type exchange-correlation functions, respectively33,34. The 3 × 1 × 1 and 1 × 4 × 1 k-point meshes were adopted for the system of the SAMs aligned along armchair and zigzag directions, respectively. All the structures were released until the force acting on each atom was less than 0.01 eV/Å, with the energy convergence of 10-4 eV. The plane wave cut-off energy was set as 400 eV. A vacuum layer with a thickness greater than 15 Å was constructed to avoid the interactions between two neighboring images. Besides, all calculations include spin-polarization and DFT-D3 was chosen to correct the vdW interaction between graphene and SAMs35.
Characterization of GSAMs by cryo-EM
The aforementioned TEM grid coated by suspended GSAMs was directly inserted into liquid nitrogen (−196 °C), and uploaded into a Thermo Fisher Scientific Titan Krios (300 kV) TEM, which is equipped with a direct electron detector K3 (Gatan, Inc.) and a GIF-Quantum energy filter. The suspended GSAMs membrane was imaged at a nominal magnification of 165,000, with a calibrated pixel size of 0.5141 Å. The total irradiation dose is 50 e-/Å2.
Binding energy between the graphene and the SAMs
The periodic models of two surfactant molecules on graphene lattice were built to calculate their binding energies (\({E}_{b}\)) by the following equation,
in which \({E}_{{SAM}-{Gr}}\), \({E}_{{SAM}}\), \({E}_{{Gr}}\), and \({N}_{{Gr}}\) are the total energy of SAMs adsorbed on monolayer graphene, the energy of SAMs, the energy of monolayer graphene and the number of carbon atoms in monolayer graphene, respectively. Finally, the binding energy between SAMs and graphene arranged along the zigzag lattice direction was calculated to be −199.0 mJ/m2.
Cryo-EM sample preparation
Streptavidin was purchased from New England Biolabs (Catalog: N7021S). To purify the 20S proteasome, its β subunit was His-tagged and expressed in Escherichia coli cells. The protein was then purified using nickel column (GE Healthcare) and stored at 0.6 mg/mL for subsequent usage18. ACE2 was purified from HEK293T cells using a C-terminal six-histidine tag and a nickel affinity column (GE Healthcare). SARS-CoV-2 RBD, tagged with a C-terminal six-histidine tag, was purified from HEK293F cells and mixed with ACE2 at a molar ratio of 1:1.5 to form the ACE2-RBD complex. The Spike protein, tagged with a C-terminal Flag, was purified from HEK293F cells36. All three samples were diluted to 0.25 mg/mL for cryo-EM specimen preparation. CHAPSO (3-(3-cholamidopropyl-dimethylammonio)-2-hydroxy-1-propanesulfonate) was purchased from Sigma-Aldrich (Catalog: C3649-500MG), and DDM (dodecyl-β-D-maltoside) was purchased from Inalco Pharmaceuticals (Catalog: 69227-93-6).
To prepare the cryo-EM specimen, 3 μL sample was pipetted onto the glow-discharged (Harrick Plasma, low-power mode for 15 seconds) graphene grid or GSAMs grid, which was then blotted for 0.5 s by filter papers with the force −2 in a Vitrobot (Thermo Fisher Scientific) under 8 °C and 100% humidity. These grids were then directly plunged into liquid ethane and transferred to liquid nitrogen for storage.
To prepare the cryo-EM specimen in the presence of surfactants, we added CHAPSO or DDM into the sample solution at final concentrations of 8 mM and 0.17 mM and then applied the sample solution onto glow-discharged EM grids (Quantifoil, 200 mesh, 1.2/1.3). The grids were blotted for 2.5 s with the force −2 in a Vitrobot (Thermo Fisher Scientific) under 8 °C and 100% humidity. After blotting, the grids were frozen by liquid ethane and stored in liquid nitrogen.
Single-particle cryo-EM data acquisition and analysis
For 20S proteasome on GSAMs grid, ACE2-RBD and Spike on both GSAMs grid and pristine graphene grid, cryo-EM datasets were automatically collected by EPU software on a Thermo Fisher Scientific Titan Krios G4 (300 kV) TEM equipped with a direct electron detector Falcon 4 (Thermo Fisher Scientific), with the defocus from −1.5 to −1.8 μm and pixel size of 0.86 Å, and we collected 1513, 1445, 3810, 921, and 4352 micrographs for aforementioned samples, respectively. For ACE2 and streptavidin on GSAMs grids, 2448 and 2144 micrographs were collected via a Thermo Fisher Scientific Titan Krios G3i (300 kV) TEM equipped with a direct electron detector K3 (Gatan, Inc.) and GIF-Quantum energy filter by AutoEMation software37, with the defocus from −1.2 to −1.5 μm and pixel size of 0.5191 Å. For ACE2 on graphene grid, 5576 micrographs were collected via a Thermo Fisher Scientific Titan Krios (300 kV) TEM equipped with a direct electron detector K3 (Gatan, Inc.) and GIF-Quantum energy filter by AutoEMation software, with the defocus from −1.2 to −1.5 μm and pixel size of 0.5141 Å. All of the micrographs were fractionated to 32 frames with a total irradiation dose of ~50 e-/Å2, which were then motion-corrected by MotionCor238 for following structural reconstruction. More details were summarized in Supplementary Table 1.
The micrographs were then imported into CryoSPARC39 to perform patch CTF estimation and particle picking. Several rounds of 2D and 3D classification were applied to exclude bad particles. A final Non-uniform Refinement with local CTF refinement was used for high-resolution structure determination. Angular distribution plots were generated by RELION40. To build the atomic models, 6J6K, 7W9I, 7W92, and 3J9I from PDB databank were fetched to rigid-body fit into the maps of streptavidin, ACE2 (ACE2-RBD), Spike, and 20S proteasome in Chimera41, respectively, and then real-space refined in PHENIX42. All these cryo-EM data collection, refinement and validation statistics are summarized in Supplementary Table 1. Efficiency of angular distribution was calculated by cryo-EF28.
To avoid the potential biases when comparing the orientation distribution from different datasets, we performed the reconstruction of experimental and control groups with identical procedures, parameters, and selection criteria in CryoSPARC39. During 3D reconstruction, the same reference was used. The final number of particles used for plotting Euler angle distribution was also identical.
Cryo-ET data acquisition and analysis
The cryo-ET tilt series were acquired from +60 to −60° at the step of 3°, on a Thermo Fisher Scientific Titan Krios (300 kV) TEM equipped with a K3 camera (Gatan, Inc.) using SerialEM. The imaging defocus was set to −5 μm with a pixel size of 1.25 Å, and each tilt was fractionated to 8 frames with a total dose of ~3 e-/Å2. The beam-induced motion was corrected by MotionCor238, and then the tilt series were imported into IMOD43 for alignment and tomogram reconstruction, followed by denoising via IsoNet44. The positions of 20S proteasome were marked by template matching, as well as visual interpretation.
Statistics and reproducibility
The experiments in Figs. 2b, 2e, 2f, and 3g–i were repeated for more than 3 times independently with similar results, including the STM characterizations of GSAMs, SEM imaging of suspended GSAMs membranes, cryo-EM characterizations of suspended GSAMs, cryo-EM imaging of 20S proteasome particles with the presences of GSAMs, CHAPSO, and DDM.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The cryo-EM density maps and coordinates of streptavidin on GSAMs, Spike on GSAMs, Spike on graphene membrane, ACE2-RBD on GSAMs, ACE2-RBD on graphene membrane, ACE2 on GSAMs, and ACE2 on graphene membrane, have been deposited into the Electron Microscopy Data Bank (EMDB) under the accession numbers EMD-34978, EMD-34975, EMD-34974, EMD-34976, EMD-34977, EMD-34979, and EMD-34980, and into the Protein Data Bank (PDB) under the accession numbers 8HRM, 8HRJ, 8HRI, 8HRK, 8HRL, 8HRN, and 8HRU, respectively. The raw data of streptavidin and ACE2 on GSAMs have been deposited into the EMPIAR, with the corresponding accession codes EMPIAR-12039 and EMPIAR-12040, respectively. Any additional data supporting the results in this work are available from the corresponding authors upon reasonable requests. The source data underlying Figs. 2e, 3b, 3e, 4k, 4l, 5c and Supplementary Figs. 3b, 9b are provided as a Source data file. Source data are provided with this paper.
References
Han, B. G., Avila-Sakar, A., Remis, J. & Glaeser, R. M. Challenges in making ideal cryo-EM samples. Curr. Opin. Struc. Biol. 81, 102646 (2023).
Glaeser, R. M. Preparing better samples for cryo–electron microscopy: biochemical challenges do not end with isolation and purification. Annu. Rev. Biochem. 90, 451–474 (2021).
Weissenberger, G., Henderikx, R. J. M. & Peters, P. J. Understanding the invisible hands of sample preparation for cryo-EM. Nat. Methods 18, 463 (2021).
Naydenova, K., Jia, P. P. & Russo, C. J. Cryo-EM with sub-1 angstrom specimen movement. Science 370, 223 (2020).
Taylor, K. A. & Glaeser, R. M. Retrospective on the early development of cryoelectron microscopy of macromolecules and a prospective on opportunities for the future. J. Struct. Biol. 163, 214 (2008).
Noble, A. J. et al. Routine single particle cryoEM sample and grid characterization by tomography. Elife 7, e34257 (2018).
Liu, N. & Wang, H. W. Better Cryo-EM specimen preparation: how to deal with the air-water. Interface? J. Mol. Biol. 435, 167926 (2023).
Basanta, B., Hirschi, M. M., Grotjahn, D. A. & Lander, G. C. A case for glycerol as an acceptable additive for single-particle cryoEM samples. Acta Crystallogr. D. 78, 124 (2022).
Chen, S. X., Li, J. D., Vinothkumar, K. R. & Henderson, R. Interaction of human erythrocyte catalase with airwater interface in cryoEM. Microsc.-Jpn 71, i51 (2022).
Carragher, B. et al. Current outcomes when optimizing ‘standard’ sample preparation for single-particle cryo-EM. J. Microsc-Oxf. 276, 39 (2019).
Frederik, P. M., Stuart, M. C. A., Bomans, P. H. H. & Busing, W. M. Phospholipid, natures own slide and cover slip for cryo-electron microscopy. J. Microsc-Oxf. 153, 81 (1989).
Chen, J., Noble, A. J., Kang, J. Y. & Darst, S. A. Eliminating effects of particle adsorption to the air/water interface in single-particle cryo-electron microscopy: bacterial RNA polymerase and CHAPSO. J. Struct. Biol.-X 1, 100005 (2019).
Fan, H. C. et al. A cryo-electron microscopy support film formed by 2D crystals of hydrophobin HFBI. Nat. Commun. 12, 7257 (2021).
Han, B. G. et al. Long shelf-life streptavidin support-films suitable for electron microscopy of biological macromolecules. J. Struct. Biol. 195, 238 (2016).
Wang, L. G., Ounjai, P. & Sigworth, F. J. Streptavidin crystals as nanostructured supports and image-calibration references for cryo-EM data collection. J. Struct. Biol. 164, 190 (2008).
Naydenova, K., Peet, M. J. & Russo, C. J. Multifunctional graphene supports for electron cryomicroscopy. P. Natl Acad. Sci. USA 116, 11718 (2019).
Han, Y. M. et al. High-yield monolayer graphene grids for near-atomic resolution cryoelectron microscopy. Proc. Natl Acad. Sci. USA 117, 1009 (2020).
Liu, N. et al. Bioactive functionalized monolayer graphene for high-resolution cryo-electron microscopy. J. Am. Chem. Soc. 141, 4016 (2019).
D’Imprima, E. et al. Protein denaturation at the air-water interface and how to prevent it. Elife 8, e42747 (2019).
Palovcak, E. et al. A simple and robust procedure for preparing graphene-oxide cryo-EM grids. J. Struct. Biol. 204, 80 (2018).
Wang, F. et al. General and robust covalently linked graphene oxide affinity grids for high-resolution cryo-EM. Proc. Natl Acad. Sci. USA 117, 24269 (2020).
Zheng, L. M. et al. Uniform thin ice on ultraflat graphene for high-resolution cryo-EM. Nat. Methods 20, 123 (2023).
Lin, Y. C. et al. Graphene annealing: how clean can it be? Nano Lett. 12, 414 (2012).
Zhang, Z. K. et al. Rosin-enabled ultraclean and damage-free transfer of graphene for large-area flexible organic light-emitting diodes. Nat. Commun. 8, 14560 (2017).
Leong, W. S. et al. Paraffin-enabled graphene transfer. Nat. Commun. 10, 867 (2019).
Snijder, J. et al. Vitrification after multiple rounds of sample application and blotting improves particle density on cryo-electron microscopy grids. J. Struct. Biol. 198, 38 (2017).
Zheng, L. M. et al. Robust ultraclean atomically thin membranes for atomic-resolution electron microscopy. Nat. Commun. 11, 541 (2020).
Naydenova, K. & Russo, C. J. Measuring the effects of particle orientation to improve the efficiency of electron cryomicroscopy. Nat. Commun. 8, 629 (2017).
Sun, L. Z. et al. Toward epitaxial growth of misorientation-free graphene on Cu(111) foils. ACS Nano 16, 285 (2022).
Zhang, J. C. et al. Clean transfer of large graphene single crystals for high-intactness suspended membranes and liquid cells. Adv. Mater. 29, 1700639 (2017).
Kresse, G. & Furthmuller, J. Efficiency of ab-initio total energy calculations for metals and semiconductors using a plane-wave basis set. Comp. Mater. Sci. 6, 15 (1996).
Kresse, G. & Hafner, J. Ab-initio molecular-dynamics for open-shell transition-metals. Phys. Rev. B 48, 13115 (1993).
Perdew, J. P., Burke, K. & Ernzerhof, M. Generalized gradient approximation made simple. Phys. Rev. Lett. 77, 3865 (1996).
Kresse, G. & Joubert, D. From ultrasoft pseudopotentials to the projector augmented-wave method. Phys. Rev. B 59, 1758 (1999).
Grimme, S., Antony, J., Ehrlich, S. & Krieg, H. A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J. Chem. Phys. 132, 154104 (2010).
Niu, S. et al. Molecular basis of cross-species ACE2 interactions with SARS-CoV-2-like viruses of pangolin origin (vol 40, e107786, 2021). EMBO J. 41, e109962 (2022).
Lei, J. L. & Frank, J. Automated acquisition of cryo-electron micrographs for single particle reconstruction on an FEI Tecnai electron microscope. J. Struct. Biol. 150, 69 (2005).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331 (2017).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290 (2017).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, e42166 (2018).
Pettersen, E. F. et al. UCSF chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605 (2004).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. 66, 213 (2010).
Kremer, J. R., Mastronarde, D. N. & McIntosh, J. R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71 (1996).
Liu, Y. T. et al. Isotropic reconstruction for electron tomography with deep learning. Nat. Commun. 13, 6482 (2022).
Acknowledgements
We are grateful to the Electron Microscopy Laboratory and Molecular Materials and Nanofabrication Laboratory (MMNL) in the College of Chemistry at Peking University for the use of instruments. The work is supported by the National Natural Science Foundation of China (52021006, T2188101 to H.L.P. and Z.F.L; 31825009 to H.-W.W.; 32200981 to N.L.), Beijing National Laboratory for Molecular Sciences (BNLMS-CXTD-202001), the New Cornerstone Science Foundation through the XPLORER PRIZE (to H.-W.W. and H.L.P.). The authors acknowledge the supports of High-Talent Support from Shenzhen Institute of Advanced Technology (SIAT-SE3G0991010, 2023) and startup grant from Shenzhen Institute of Advanced Technology (to F.D.). N.L. was supported by China Postdoctoral Science Foundation (2021M701919).
Author information
Authors and Affiliations
Contributions
L.Z., N.L., H.-W.W. and H.P. conceived the project and designed the experiments. L.Z. and X.Y.G. prepared the GSAMs grid and graphene grid. N.L., J.X., H.C., X.J. and F.M. carried out the single-particle cryo-EM characterizations and analysis. N.L. and L.Z. carried out the cryo-EM characterizations and analysis of GSAMs. C.Z., W.W. and F.D. performed the DFT calculations. W.G., X.Z. and K.W. performed the STM characterizations. W.S. performed the experiment in Supplementary Movie. L.S., B.C. and Z.L. prepared the graphene films on the copper foils. L.Z., N.L., H.-W.W. and H.P. wrote the manuscript. All authors discussed the results and commented on the manuscript. All authors have given approval to the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors, Liming Zheng, Xiaoyin Gao, and Hailin Peng have filed the Chinese patent applications (ZL202210107288.7, ZL201911343879.9), covering the procedures of preparing the free-standing GSAMs superstructure, suspended GSAMs membranes, and suspended graphene membranes. Beijing Graphene Institute contributes to the growth of graphene films. Shuimu Biosciences provides the services of cryo-EM imaging and protein structural determination. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Oliver Biggs Clarke, Alex Noble and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zheng, L., Xu, J., Wang, W. et al. Self-assembled superstructure alleviates air-water interface effect in cryo-EM. Nat Commun 15, 7300 (2024). https://doi.org/10.1038/s41467-024-51696-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-51696-w
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.