Abstract
Error-free replication of DNA is essential for life. Despite the proofreading capability of several polymerases, intrinsic polymerase fidelity is in general much higher than what base-pairing energies can provide. Although researchers have investigated this long-standing question with kinetics, structural determination, and computational simulations, the structural factors that dictate polymerase fidelity are not fully resolved. Time-resolved crystallography has elucidated correct nucleotide incorporation and established a three-metal-ion-dependent catalytic mechanism for polymerases. Using X-ray time-resolved crystallography, we visualize the complete DNA misincorporation process catalyzed by DNA polymerase η. The resulting molecular snapshots suggest primer 3´-OH alignment mediated by A-site metal ion binding is the key step in substrate discrimination. Moreover, we observe that C-site metal ion binding preceded the nucleotidyl transfer reaction and demonstrate that the C-site metal ion is strictly required for misincorporation. Our results highlight the essential but separate roles of the three metal ions in DNA synthesis.
Similar content being viewed by others
Introduction
DNA polymerases catalyze the essential biological process of DNA replication. Based on their amino acid sequences, these vital enzymes have been classified into A, B, C, D, X, Y, reverse transcriptase (RT), and primase and polymerase (PrimPol) families1. Despite their various biological roles and subunit compositions, all DNA polymerases contain similar active sites and catalyze the metal-ion dependent nucleotidyl transfer reaction via similar kinetic pathways. Moreover, most DNA polymerases resemble a right hand architecture with the active site residing in the palm domain, a thumb domain that binds the primer:template DNA, and a finger domain that interacts with the nascent base-pair. All polymerase active sites contain two to three conserved acidic residues that coordinate two metal ions (Fig. 1a). The A-site metal ion (Me2+A) bridges the primer 3′-OH and the substrate α-phosphate to promote deprotonation and nucleophilic attack, while the B-site metal ion (Me2+B) stabilizes the triphosphate motif of the incoming deoxynucleotide triphosphate (dNTP). Drawn from many early crystal structures, it was believed that two metal ions were required and sufficient for catalysis;2 however, recent time-resolved crystallography experiments revealed that polymerases acquired a third metal ion (Me2+C) during catalysis, suggesting a three-metal-ion dependent mechanism3,4,5,6 (Fig. 1a). The observed Me2+C bound between the α- and β-phosphates and may have driven α-β-phosphate bond breakage3. Similarly, a two-metal-ion dependent catalysis mechanism has also been proposed for many nucleases; however, additional divalent metal ions were observed in RNaseH7 and Endonuclease V8 with time-resolved crystallography. In addition, three metal-ion dependent catalysis was also observed in non-nucleic acid enzymes, fructose-1,6-bisphosphatase9 and type I terpenoid synthases10, suggesting a widespread mechanism in promoting phosphorus-oxygen bond breakage and formation.
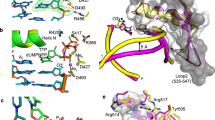
a Scheme of three-metal-ion dependent enzyme catalysis and transition state stabilization. Divalent metal ions optimally bind to their ligands with bond lengths of 2.0–2.2 Å at 90°. b Geometries of dGTP:dC Watson Crick base-pair and dGTP:dT wobble base-pair. c DNA polymerase fidelity in the presence of Mg2+ versus Mn2+ based on steady-state kinetics. The bars represent the mean of duplicate measurements for the catalytic efficiencies (kcat/KM) for incorporation of dATP (blue) and dGTP (red) opposite dT for Pol η with Mg2+ and Mn2+(Supplementary Fig. 1). The errors bars represent the standard deviation for the measurements. The distance between the respective catalytic efficiencies is a measure of discrimination. d Structure of Pol η ground state complexed with Ca2+. The 2Fo-Fc map for the primer up conformation, Me2+A, Me2+B, dGTP, and catalytic residues (blue) was contoured at 2 σ (σ values represent r.m.s. density values). The Fo-Fc omit map for the down conformation of the primer (red) was contoured at 3 σ. e dGTP:dT wobble base-pairing (red and dark cyan), dATP (pink), and primer (yellow) in the Pol η ground state. The 2Fo-Fc map for dGTP, template dT, two conformations of R61, and Q38 (blue) was contoured at 1.8 σ.
During DNA replication, polymerases are faced with the task of incorporating the correct nucleotide from a pool of nearly identical-looking dNTPs dictated by the template bases. Structural studies with various polymerases have suggested that base tautomerization11,12, solvation of the active site13, and non-Watson-Crick geometries such as Hoogsteen14, syn-trans15, and wobble base15,16 geometries (Fig. 1b) help to stabilize mismatches and reduce steric clashes. Intrinsic fidelity in most polymerases, however, is much higher than what can be provided by the difference in base-pairing energy1,17,18. Pre-steady-state kinetics have suggested that a rate-limiting conformational step proceeds catalysis and might be the key in substrate discrimination19,20. Based on early structural work, an open-to-closed transition occurs in the finger domain in most replicative polymerases following dNTP binding21. It was thus proposed that finger domain closure was the key step during misincorporation19,20,22. This finger domain movement, however, is absent in Y-family polymerases and was too fast to be rate-limiting in neither correct nor incorrect nucleotide incorporation1,23,24,25. Subsequently, biophysical studies of X-family DNA polymerase β (Pol β) and Y-family Dpo4 polymerase have suggested that a local “micro” conformational change within the finger domain might play a role in polymerase fidelity26,27,28. On the other hand, work from Tsai, Johnson, Goodman, and co-workers argued that a rate-limiting conformational change was unnecessary to explain the kinetic data and that a kinetic balance between incoming dNTP release and nucleotidyl transfer could account for polymerase fidelity25,29,30,31. Other fluorescent studies of Pol β have suggested that the rate-limiting step requires a functional primer32 and have proposed that the rate-limiting step is dependent on primer alignment33. Time-resolved crystallography can potentially reveal information about the dynamic catalytic process at atomic resolution4,5,6,7,34,35,36. Structural snapshots of Pol β during misincorporation have revealed that an incoming dATP formed a buckle base-pair with a template dG and that the primer terminus and α-phosphate in both the substrate and product states were misaligned5. However, intermediate structures right before and right after the chemistry step were not captured and how the substrates were aligned prior to nucleotidyl attack remains elusive. Moreover, in contrast with correct nucleotide incorporation5,34, the Me2+C was not captured during the misincorporation process.
DNA polymerase η (Pol η) is a Y-family polymerase involved in DNA translesion synthesis against UV-induced lesions37. Individuals with mutations on the Pol η gene develop a hypersensitivity to UV-radiation and often have a predisposition for xeroderma pigmentosum and skin cancer38,39. In addition, Pol η is involved in antibody generation as well as prevents tumor cellular arrest and confers drug resistance40,41,42,43,44,45. The Pol η time-resolved crystallographic model system has been used extensively to investigate the catalytic mechanism of DNA polymerases4,34,36,46,47. Utilizing this Pol η system, the reaction process of correct nucleotide incorporation has been captured and a transiently bound Me2+C has been proven essential for catalysis4,34. Static structures of Pol η incorporating incorrect nucleotides have also been reported48. The incorrect substrate dGMPNPP formed a wobble base with the template dT, which was stabilized by protein sidechains within the Pol η active site. However, the reaction process was not resolved and the key structural determinant for fidelity remained unclear. In this study, we determine 34 atomic-resolution structures from time-resolved X-ray crystallography and elucidate the entire misincorporation process by Pol η with Mg2+ and Mn2+. Our results suggest that primer 3´-OH-substrate alignment by the Me2+A is the key step in substrate discrimination. We also highlight Mn2+’s superior ability in aligning the 3′-OH, explaining its error-prone nature compared to Mg2+. Furthermore, we show that the Me2+C binds before the reaction, and support its essential role in promoting catalysis.
Results
Biochemical assays
Pol η has previously been shown to be more error-prone on DNA substrates that contain the WA motif (5′-AA or 5′-TA) on the primer termini48. We reasoned that the error-prone WA substrate would be useful in facilitating mechanistic dissections of misincorporation. Steady-state kinetic assays were conducted to confirm Pol η’s discrimination against incorrect substrate insertion on a DNA substrate that contained a WA motif at the primer termini. With Mg2+, the steady-state efficiencies indicated that the incorrect substate was discriminated by ~52-fold against the correct substrate, similar to the previous report48 (Fig. 1c and Supplementary Fig. 1). It is widely known that Mn2+ can support polymerase catalysis and stimulate misincorporation. Therefore, we characterized the efficiency of correct and incorrect substrate insertion by Pol η with Mn2+. The kinetic assays indicated that correct nucleotide incorporation efficiency was roughly similar in the presence of Mg2+ or Mn2+, but Mn2+ strongly increased incorrect nucleotide incorporation efficiency. Incorporation of dGTP against dT in the presence of Mn2+ was only four-fold less efficient than the incorporation of dATP across dT, resulting in a ~13-fold decrease in discrimination compared to that with Mg2+. These results confirmed that Pol η is error-prone and that Mn2+ significantly reduces substrate discrimination.
dGTP binds as a wobble base in Pol η’s active site
Following our previous protocol49, we grew Pol η crystals complexed with a DNA substrate that contained a WA motif at the primer termini and a dT as the nascent template, incorrect dGTP substrate, and the inhibitory divalent metal ion, Ca2+, at a low pH of 6.0 (Supplementary Fig. 2a). The overall structure including the finger domain looked identical to that of correct nucleotide incorporation, having an r.m.s.d. of 0.2 Å (Supplementary Fig. 3). Within the active site, the incorrect dGTP was assigned with 80% occupancy according to its density. In the ground state, t = 0 s, the Me2+A site was occupied at 30% by a monovalent K1+, and the Me2+B site was occupied at 80% by a divalent Ca2+ (Fig. 1d). This structure was similar to the ground state during correct nucleotide incorporation (Supplementary Fig. 4a), in which the Me2+A site remained partially occupied by K1+ while the Me2+B site was occupied by Ca2+ at 100%. In our mismatch structure, the majority of the R61 side chain was rotated away from the triphosphate group of dGTP, and the primer end existed in two conformations, up and down like in the static structure with a nonhydrolyzable dGMPNPP and two Mg2+ ions48 (Supplementary Fig. 4c). In the up conformation, the primer 3′-OH was located 3.7 Å above the substrate α-phosphate while the sugar moiety interacted with R61. In the down conformation, the 3′-OH existed 3.6 Å away from the K1+ and 3.3 Å away from S113 (Fig. 1d and Supplementary Fig. 4e). Assigning 15% of the 3′-OH in the down conformation eliminated any significant Fo-Fc peaks around the primer terminus. In contrast, during correct nucleotide incorporation, all of the primer was in the down conformation stabilized by S113 (Supplementary Figs. 4a and f). Regarding base-pair geometry of our mismatch structure, the incorrect substrate dGTP and template dT formed a wobble base-pair, in which the base of the incoming dGTP shifted 1.6 Å towards the minor groove while the template dT shifted 0.9 Å towards the major groove, compared to the ground state structure with the correct substrate dATP (Fig. 1e and Supplementary Figs. 4b and d). Hydrogen bonds between dGTP and the protein sidechains Q38 and R61 possibly stabilized the wobble base-pair. Furthermore, due to the wobble base geometry, there was a reduction in pi-stacking interactions between the incoming dGTP and primer aromatic base.
In crystallo observation of misincorporation by Pol η in the presence of Mg2+
To start the reaction, we equilibrated Pol η crystals at pH 7.0 and then soaked the crystals in reaction buffer containing Mg2+ or Mn2+ (Supplementary Fig. 1a). After soaking the Pol η crystals in 1 mM Mg2+ reaction buffer for 40 s, K1+ dissociated from the Me2+A site, and Ca2+ started to dissociate from the Me2+B site, leaving 60% and 50% of Mg2+ in the Me2+A and Me2+B sites, respectively (Fig. 2a). Binding of the Mg2+A induced partial primer 3′-OH alignment. Around 35% of the primer end resided in the aligned down conformation, with the 3′-OH existing 2.5 Å from the Mg2+A, 0.5 Å longer than the perfect coordination distance for Mg2+50. The 3′-OH was ~175° in-line and 3.6 Å away from the α-phosphate, which may be too far for initiating a nucleophilic attack. In contrast, for the Pol η crystals with the correct substrate that were soaked in the same condition for the same reaction time, all of the primer end was found in the down conformation, existing 2.2 Å away from the Mg2+A as well as 3.2 Å away and ~174° in-line from the α-phosphate.
a–c Structures of Pol η during in crystallo catalysis after 1 mM Mg2+ soaking for 40 s (a), 80 s (b), and 140 s (c). The 2Fo-Fc map for the primer up conformation (blue) was contoured at 1.2 σ. The 2Fo-Fc map for the Me2+A, Me2+B, dGTP, and catalytic residues (blue) was contoured at 2 σ in (a–c). The Fo-Fc omit map for the down conformation of the primer (green) was contoured at 3.3 σ in (a–c). The Fo-Fc omit map for all the Mg2+C, and newly formed bond (red) was contoured at 4 σ. d Timescale of Mg2+A, Mg2+C binding and reaction product in crystallo with Mg2+. e, f Observation of the Mg2+C after 1 mM Mg2+ soaking for 80 s (e) and 140 s (f). g Correlation (R2) between the newly formed bond and the Mg2+C binding. d, g The data points represent singular measurements for r.m.s. density values of electron density. In (e, f), the 2Fo-Fc map for the water molecules that were coordinated by the Mg2+C was contoured at 0.7 σ. The Fo-Fc omit map for the Mg2+C, new bond, and Mg2+A and Mg2+B was contoured at 4 σ. h Alternative product state of Pol η in crystallo with 1 mM Mg2+ at 300 s. The 2Fo-Fc map was contoured at 1 σ.
We next tracked the electron density of the product phosphate and the Me2+C during catalysis. After soaking for 80 s in 1 mM Mg2+, the primer’s position moved 0.2 Å closer to the α-phosphate (Fig. 2b). Surprisingly, the Mg2+C was observed while no density was detected for the converted product. The Mg2+C at this timepoint was 2.5 Å and 2.6 Å away from the pro-Sp phosphate oxygen and α-β-phosphate bridging oxygen, respectively, with a bond-angle of 70° (Fig. 2e). In addition, four water molecules completed the octahedral coordination for the Mg2+C. We assigned the Mg2+C with an occupancy of 0.15, at which the B-factor of the surrounding atoms was similar to that of the Mg2+C. After soaking for 140 s, compared to the 80 s structure, the primer moved 0.2 Å closer to the substrate α-phosphate (3.2 Å away) (Fig. 2c) and the Mg2+C moved 0.5 Å closer to the product oxygen (2.2 Å and 2.3 Å away from the product oxygen and pyrophosphate (PPi) oxygen respectively) (Fig. 2f and Supplementary Fig. 5a). In conjunction with the shift of the Mg2+C, density for the product phosphate appeared. With a longer incubation time of 250 s, densities corresponding to both the new bond and the Mg2+C increased, and the Mg2+C remained well coordinated by the product oxygen and PPi oxygen (Fig. 2d). Just like in correct nucleotide incorporation, the Mg2+C density correlated well with the density of the product phosphate (R2 = 0.98) (Fig. 2g). It is important to note that the first detected density for the Mg2+C did not cross the origin in the Mg2+C-bond formation correlation plot, as it appeared before any product was detected.
Previous studies with Pol η4,48, Pol β5,6, Bacillus51, and Geobacillus35 DNA Pol I have revealed that the incorporated nucleotide moves to the primer position after nucleotidyl transfer and PPi dissociation. For three crystals soaked in 1 mM Mg2+ reaction buffer for 300 s, moderate density was detected in the spacious region of Pol η’s active site away from the catalytic residues. The product state translocated almost 20 Å and flipped out of the active site to reside in close proximity to an alanine residue in the neighboring unit cell (Fig. 2h). In place of the product state, water molecules were detected within the active site.
In crystallo observation of misincorporation by Pol η in the presence of Mn2+
We also investigated the misincorporation process supported by error-prone Mn2+. After soaking in 10 mM Mn2+ for 30 s, Mn2+ occupied 65% and 80% at the Me2+A and Me2+B sites, respectively. At this point, 40% of the primer end existed in the down conformation, with the 3′-OH 2.5 Å away from the Mn2+A and 3.4 Å away to the α-phosphate (Fig. 3a). The distances between the Me2+A and the 3′-OH were greater than the ideal coordination distances with both Mg2+ and Mn2+50, indicating that 3′-OH alignment during misincorporation was unfavorable.
a, b Structures of Pol η during in crystallo catalysis after 10 mM Mn2+ soaking for 30 s (a) and 120 s (b). The 2Fo-Fc map for the primer up conformation (blue) was contoured at 1.2 σ. The 2Fo-Fc map for the Me2+A, Me2+B, dGTP, and catalytic residues (blue) were contoured at 2.5 σ in (a–c). The Fo-Fc omit map for the down conformation of the primer (green) was contoured at 3.3 σ in (a–c). The Fo-Fc omit map for all the Mn2+C, and newly formed bond (red) was contoured at 4 σ. cTimescale of Mn2+A, Mn2+C binding and reaction product in crystallo with Mn2+. d, e Observation of the Mn2+C after 10 mM Mn2+ soaking for 30 s (d) and 120 s (e). f Correlation (R2) between the newly formed bond and the Mn2+C binding. In d, e, the 2Fo-Fc map for the water molecules that were coordinated by the Mn2+C was contoured at 0.7 σ. The Fo-Fc omit map for the Mn2+C, new bond, and Mn2+A and Mn2+B was contoured at 4 σ. c, f The data points represent singular measurements for r.m.s. density values of electron density. g Alternative product state of Pol η in crystallo with 10 mM Mn2+ at 300 s. The 2Fo-Fc map for the primer up conformation, primer down conformation, Me2+A, Me2+B, Me2+C, and dGTP (blue) was contoured at 2 σ. The Fo-Fc omit map for the up conformation of the product state (red) was contoured at 3 σ.
Similar to the Mg2+ time-resolved results, after 30 s of soaking in 10 mM Mn2+, the Mn2+C was observed (Fig. 3a, d). The Mn2+C was coordinated by four water molecules and the incoming dGTP, but the Mn2+C remained too far from the phosphate oxygen and bridging oxygen (2.6 Å and 2.3 Å away). We assigned the Mn2+C with an occupancy of 0.10, and the B-factor of the surrounding atoms after refinement was similar to that of the Mn2+C. Furthermore, longer soaking times of 60 s and 90 s in Mn2+ resulted in larger densities for the Mn2+C, of which were assigned with 0.13 and 0.15 occupancies, respectively, but the product density was still not observed. After 120 s of Mn2+ soaking, small amount of product was observed, and the Mn2+C was found to be 0.5 Å closer towards the phosphate oxygen, compared to the 90 s structure (Fig. 3b, e and Supplementary Fig. 5b, c). After 180 s, both Mn2+C and new bond densities grew larger, indicating more Mn2+C binding and product formation (Fig. 3c). Just like for the reaction in Mg2+, the Mn2+C density correlated well with the density of the product phosphate (R2 = 0.88) (Fig. 3f) and did not cross the origin in the Mn2+C-bond formation correlation plot. Contrasting with the flipped-out product state with Mg2+, a fraction of the product shifted 2.5 Å away from the Mn2+A and Mn2+B sites (Fig. 3g) and remained in a wobble base-pair with the template dT after 300 s soaking.
The Me2+ C is essential for misincorporation
The Me2+C had been shown to be essential for correct nucleotide incorporation in previous time-resolved reports34. Because our time-resolved results illustrated that the Me2+C appeared in the reactant states for both Mg2+ and Mn2+, we wanted to confirm if the Me2+C was essential for misincorporation. In previous experiments, the Me2+A and Me2+B binding could be separated from Me2+C binding and catalysis by soaking the Pol η crystals in low concentrations of Mn2+34. Soaking Pol η crystals in 0.5 mM Mn2+ buffer for 30 min allowed Mn2+ to saturate the Me2+A and Me2+B sites to ~80% occupancy (Fig. 4a). At this stage, the majority (50%) of the primer was aligned with the substrate. The primer 3′-OH was 3.1 Å away from the α-phosphate and the 3′-OH was 2.3 Å away from the Mn2+A. Yet, neither the Mn2+C nor product densities were observed. The absence of product formation implied that something essential for catalysis was missing. To test if the Mn2+C was the missing component, a two-step reaction was performed by first soaking the crystal in 0.5 mM Mn2+ for 30 min and then in reaction buffer that contained 10 mM Mn2+ (Supplementary Fig. 2b). After 30 s of soaking in 10 mM Mn2+, the Mn2+C density was detected before product formation and assigned with an occupancy of 0.10 (Fig. 4b); however, the Mn2+C was not in an ideal octahedrally coordinated complex, existing 2.7 Å and 2.3 Å away from the substrate pro-Sp oxygen and α-β-phosphate bridging oxygen, respectively. After 300 s of soaking, both the product and the Mn2+C were clearly present. The Mn2+C density was 2.3 Å away from both the phosphate oxygen and PPi oxygen, forming an octahedrally coordinated complex (Fig. 4c and Supplementary Fig. 5d).
Structures of Pol η in crystallo after 0.5 mM Mn2+ soaking (a) and subsequent 10 mM Mn2+ soaking for 30 s (b) and 300 s (c). The 2Fo-Fc map for all the water molecules coordinating the Me2+C was contoured at 0.7 σ. The Fo-Fc omit map for all Mn2+ ions and new bond density (red) was contoured at 3 σ.
Correlation of Me2+ A binding with primer 3′-OH-substrate alignment
Our time-resolved results suggested that primer 3′-OH alignment and Me2+A metal binding were key steps in misincorporation. However, due to conformational changes associated with product formation, it was difficult to confidently correlate primer 3′-OH alignment and Me2+A binding. To further investigate the role of the Me2+A, we used a nonhydrolyzable analog of the incorrect substrate dGMPNPP and titrated the Me2+A in crystallo with different concentrations of Mg2+ or Mn2+ (Supplementary Fig. 2c). Pol η crystals complexed with dGMPNPP were soaked in Mg2+ reaction buffer ranging from 0.025 to 2.0 mM Mg2+ for 10 min. We monitored the density of the Me2+A and the primer in the aligned conformation. Although the Mg2+A bound with a KD of 0.03 mM, rendering an omit Fo-Fc map failed to produce sufficient densities that could be confidently assigned with the down conformation of the primer sugar moiety for a Pol η crystal that was soaked in 2 mM Mg2+ for 600 s (Fig. 5a). Near the primer termini, only small spherical densities possibly corresponding to water molecules appeared in the void. We suspected that the occupancy of the down conformation of the primer was too low to be assigned (Fig. 5b, c). We conducted the similar protocol with Mn2+ reaction buffer ranging from 0.06 mM to 6.0 mM Mn2+ for 10 min, and observed that the Me2+A site could be fully saturated with a KD of 0.11 mM. In contrast with the in crystallo Mg2+A titration results, 30–40% of the primer 3′-OH was aligned for a Pol η crystal that was soaked in 3 mM Mn2+ for 600 s (Fig. 5d). A moderate correlation (R2 = 0.75) for the Me2+A and primer alignment was detected (Fig. 5e–f).
a Structure of Pol η:dGMPNPP with 2.0 mM Mg2+. b Plot showing Mg2+A density and omit map electron density for the down primer with Mg2+ concentration. c Correlation (R2) between the Mg2+A density and omit map electron density for the down primer. d Structure of Pol η:dGMPNPP with 3.0 mM Mn2+. e Plot showing Mn2+A density and omit map electron density for the down primer with Mn2+ concentration. f Correlation (R2) between the Mn2+A density and omit map electron density for the down primer conformation. The 2Fo-Fc map for the primer up conformation (blue) was contoured at 2 σ in (a) and 1.5 σ in (d). The Fo-Fc omit map for both the Me2+A and primer down conformation (red) were contoured at 3 σ. All data points shown in the plot were collected at t = 600 s and represent singular measurements.
Discussion
The dynamic reaction process of misincorporation
In this study, we captured intermediate states pre-, intra-, and post-reaction at atomic resolution, elucidating the reaction process of the misincorporation of dGTP across dT by Pol η (Supplementary Movie 1). The process of misincorporation after DNA binding begins with a bound Me2+B-complexed dGTP inside the polymerase active site. At this point, the majority of the primer 3′-OH exists in the up conformation not aligned with the α-phosphate of dGTP, and most of the R61 sidechain is interacting with the wobble base-pair (Fig. 1d, e). Upon Me2+A binding, part of the primer 3′-OH shifts downwards and approaches the substrate α-phosphate (Figs. 2a and 3a). Meanwhile, the Me2+C binds but remains far from the substrate pro-Sp oxygen (Figs. 2e and 3d). Downward movement of the Me2+C and nucleophilic attack then ensue with new-bond formation (Fig. 3b, e). In the presence of Mg2+, the product base flips out of the active site (Fig. 2h). This flip-out product may then inhibit primer extension after misincorporation. While in the presence of Mn2+, the product is stabilized within the active site (Fig. 3g). In similarity to correct nucleotide incorporation and other time-resolved studies, large movements in the substrate nucleotide were not observed4,5,34. Instead, intermediate snapshots revealed that primer 3′-OH alignment was greatly affected between correct and incorrect nucleotide incorporation. This could be attributed to unfavorable base stacking, stemming from the geometry of the dGTP:dT wobble base-pair48. Despite this, primer alignment mediated by Me2+A binding was shown not to be rate-limiting as revealed by the two-step reaction, in which primer alignment occurred before the second Mn2+ soaking step. Perturbation in primer alignment due to buckled base-pairing has also been captured in other polymerases such as the X-family Pol β, suggesting that 3′-OH misalignment is a common mechanism in preventing polymerase misincorporation52. Misalignment of the 3′-OH to the Me2+A and substrate might favor incorrect nucleotide dissociation versus nucleotidyl transfer and contribute to substrate discrimination, as suggested by Tsai, Johnson, Goodman, and co-workers25,29,30,31 (Fig. 6). On the other hand, the substrate was already bound inside Pol η’s active site in our crystal soaking set-up. Consequentially, conformational changes before and during substrate binding and after product release were not captured. Thus, we cannot rule out the possibility of other rate-limiting conformational steps during polymerase catalysis.
Proposed kinetic model during DNA polymerase correct nucleotide incorporation (a) and misincorporation (b). During correct nucleotide incorporation, the primer 3′-OH is well-aligned by the Me2+A, and the reaction proceeds right after Me2+C binding. In contrast, during incorrect nucleotide incorporation, due to reduced base-stacking, primer 3′-OH alignment is unfavored, which would lead to incorrect dNTP dissociation. Error-prone Mn2+A can drive 3′-OH alignment and once the Me2+C binds, catalysis ensues.
The Me2+ C appears before product formation
Since the Me2+C’s first visualization, the role of the Me2+C in polymerase catalysis has been extensively debated3,5,6. Similar to correct nucleotide incorporation, we observed that Me2+C binding was correlated with product formation (Figs. 2g and 3f) and reaction did not occur without Me2+C binding (Fig. 4). Without further details, one could interpret that the Me2+C only binds to and stabilizes the product state and shifts the equilibrium towards product formation. In addition, the Me2+C was only observed after product formation in Pol β and was missing from the mismatch intermediate structures5. Thus, it was proposed that the Me2+C only stabilized the product state and was not involved in driving the forward reaction. Conflicting results were reported from computational simulations53,54,55. Some have suggested that Me2+C binding does not lower the energy barrier required for catalysis but rather aids in pyrophosphate release53 while others have suggested that Me2+C binding does stabilize the transition state and significantly lowers the barrier56. The most important detail in addressing the debate lies in the timing of Me2+C binding. To be catalytic, the Me2+C must bind before product formation. We here, observed the Me2+C in the reactant state before any product formation (Figs. 2b, e, 3a, d, and 4b). This could be because R61 vacated the Me2+C binding site to stabilize the wobble base-pair; however previous studies have shown that the timing of Me2+C binding was unchanged in the R61A mutant versus WT Pol η34. Another reason could be that additional interactions by the primer up conformation helped to trap the Me2+C; however, the up primer 3′-OH existed ~3.5 Å away from the Me2+C, which may be too far for direct coordination. During its initial binding, Me2+C coordination was not an optimal octahedrally coordinated complex, as it existed 2.6–2.7 Å away from the substrate pro-Sp oxygen and α-β-phosphate bridging oxygen with a bond angle of 70°. Only when the Me2+C moved to its ideal coordination, did we see the product density. These results support that the Me2+C is catalytic and drives nucleotidyl transfer by stabilizing the transition state. Observation of a transiently bound metal ion within the active sites of metalloenzymes such as the Me2+C in Pol η has become more common with the advent of time-resolved crystallography. The increasing pervasiveness of such observations in Pol η4,34, Pol β5,6,57,58, and most recently, Pol μ59, RNaseH7, and Endonuclease V nucleases60, suggests that catalysis induced by transiently bound metal ions might be more universal than originally thought.
The Me2+ A is responsible for primer 3′-OH alignment and primer 3′-OH alignment is key in misincorporation
In this study, we showed that primer alignment was perturbed during misincorporation. This primer movement has largely been overlooked, as early ternary complex structures of DNA polymerases were complexed with dideoxynucleosides at the primer termini in order to inhibit the reaction. Without a functional 3′-OH group, many of these structures failed to capture the Me2+A or instead showed drastic changes in its position15,61. Nevertheless, the Me2+A has been thought to bridge the primer 3′-OH with the substrate α-phosphate into phosphoryl-transfer proximity and decrease the pKa of the primer 3′-OH group. Improper 3′-OH-Me2+A alignment was first captured in Pol β52 with nonhydrolyzable dNTP analogs, which revealed significant reorganizations in primer alignment. In 2012, the polymerase reaction for correct nucleotide incorporation was observed in crystallo4. Significant movement of the primer 3′-OH conformation and sugar pucker together with the binding of a water molecule proceeded nucleotidyl transfer4,34. For incorrect nucleotide incorporation, primer misalignment was observed in the time-resolved studies of Pol β, but intermediate structures that led to primer alignment were not captured5. Perturbed 3′-OH alignment was also observed in the extension of phenanthriplatin DNA damage as well as bypass of 8,5′-cyclo-2′-deoxyadenosine lesions62,63. Here, we captured primer alignment and correlated it with Me2+A binding (Fig. 5e, f). In the time-resolved results, the Me2+A was not in its ideal octahedral geometry and its bond distance was far, suggesting major barriers in aligning the 3′-OH. Mn2+ was shown to be more optimal than Mg2+ at aligning the primer, explaining why Mn2+ is error-prone in polymerase catalysis. In addition, our results suggested that the nonhydrolyzable analog dGMPNPP, although similar, is not identical to dGTP in promoting 3′-OH alignment. This is possibly due to the different electrostatic nature stemming from the nitrogen atom. Our studies again highlighted the power of observing the catalytic process with native substrates through time-resolved crystallography.
Methods
Protein expression and purification
Wild-type human polymerase η (Pol η) (residues 1–432) was cloned into a modified pET28p vector with a N-terminal 6-histidine tag and a PreScission Protease cleavage site as described47. For protein expression, this Pol η plasmid was transformed into BL21 DE3 E. coli cells. When the optical density of the E. coli cells reached 0.8, isopropyl ß-D-1-thiogalactopyranoside (IPTG) was added to a final concentration of 1 µM IPTG. After 20 h. (16 °C) of induction, the cell paste was collected via centrifugation and re-suspended in a buffer that contained 20 mM Tris (pH 7.5), 1 M NaCl, 20 mM imidazole, and 5 mM ß-mercaptoethanol (BME). After sonification, Pol η was loaded onto a HisTrap HP column (GE Healthcare), which was pre-equilibrated with a buffer that contained 20 mM Tris (pH 7.5), 1 M NaCl, 20 mM imidazole, and 5 mM BME. The column was washed with 300 mL of buffer to remove non-specific bound proteins and was eluted with buffer that contained 20 mM Tris (pH 7.5), 1 M NaCl, 300 mM imidazole, and 3 mM dithiothreitol (DTT). The eluted Pol η was incubated with PreScission Protease to cleave the N-terminal 6-histidine-tag. Afterwards, Pol η was buffer-exchanged and desalted to 20 mM 2-(N-morpholino)ethanesulfonic acid (MES) (pH 6.0), 250 mM KCl, 10% glycerol, 0.1 mM ethylenediaminetetraacetic acid (EDTA), and 3 mM DTT and was loaded onto a MonoS 10/100 column (GE Healthcare). The protein was eluted with an increasing salt (KCl) gradient. Finally, Pol η was cleaned with a Superdex 200 10/300 GL column (GE Healthcare) with a buffer that contained 20 mM Tris (pH 7.5), 450 mM KCl, and 3 mM DTT.
DNA synthesis assay
The nucleotide incorporation activity was assayed by the following: The reaction mixture contained 2.5-8 nM Pol η, 5 µM DNA, 0–400 µM dNTP (either dATP or dGTP), 100 mM KCl, 50 mM Tris (pH 7.5), 5 mM MgCl2, 3 mM DTT, 0.1 mg/mL bovine serum albumin, and 4% glycerol. The incorporation assays were executed using DNA template (5′-GAG TCA TGT TTA CGC TAG GCA C-3′) and 5′-fluorescein- labeled primer (5′-GTGCCTAGCGTAA-3′). Reactions were conducted at 37 °C for 5 min and were stopped by adding formamide quench buffer to the final concentrations of 40% formamide, 50 mM EDTA (pH 8.0), 0.1 mg/ml xylene cyanol, and 0.1 mg/ml bromophenol. After heating to 97 °C for 5 min and immediately placing on ice, reaction products were resolved on 22.5% polyacrylamide urea gels. The gels were visualized by a Sapphire Biomolecular Imager and quantified using the built-in software. Quantification of Kcat, KM, VMax and fitting and graphic representation were executed by Graph Prism. Source data of urea gels are provided as a Source Data file.
Crystallization
Pol η was concentrated to 300 µM in buffer that contained 20 mM Tris (pH 7.5), 0.45 M KCl, and 3 mM DTT. Then DNA, dGTP or dGMPNPP, and Ca2+ and low salt buffer (20 mM Tris (pH 7.5), and 3 mM DTT) were added to this polymerase solution at the molar ratio of 1:1.2:1:1 for Pol η, DNA, dGTP or dGMPNPP, and Ca2+, bringing Pol η’s concentration to 100 µM. Then after this solution was kept on ice for 10 min, more dGTP or dGMPNPP was added to a final concentration of 0.5 mM. DNA template and primer used for crystallization were 5′-CAT GAT GAC GCT-3′ and 5′-AGC GTC AT-3, respectively. All crystals were obtained using the hanging-drop vapor-diffusion method against a reservoir solution containing 0.1 M MES (pH 6.0) and 9–15% (w/v) PEG2K-MME at room temperature within 4 days.
Chemical reaction in crystallo
The crystals were first transferred and incubated in a pre-reaction buffer containing 0.1 M MES (pH 7.0, titrated by KOH), 100 µM dGTP, and 20% (w/v) PEG2K-MME for 30 min. The chemical reaction was initiated by transferring the crystals into a reaction buffer containing 0.1 M MES (pH 7.0), 20% (w/v) PEG2K-MME, and 0.5-20 mM MgCl2 or MnCl2. After incubation for a desired time period, the crystals were quickly dipped in a cryo-solution supplemented with 20% (w/v) glycerol and flash- cooled in liquid nitrogen. For the two-step in crystallo reaction, a 30 min incubation step in buffer containing 0.1 M MES (pH 7.0), 0.5 mM MnCl2, and 20% (w/v) PEG2K-MME was added before initiation of the reaction. For the A-site in crystallo titration experiment, the crystals were incubated in 0.025–6.0 mM MgCl2 or MnCl2 for 10 min.
Data collection and refinement
Diffraction data were collected at 100 K on LS-CAT beam lines 21-D-D, 21-ID-F, and 21-ID-G at the Advanced Photon Source (Argonne National Laboratory). Data were indexed in space group P61, scaled and reduced using XDS64. Isomorphous Pol η structures with Mg2+ (PDB: 4ECQ, 4ECR or 4ECV)4 and Mn2+ (PDB: 5K5G and 5L9X) were used as initial models for refinement using PHENIX65 and COOT66. Initial occupancies were assigned for the substrate, reaction product, PPi, Me2+A, Me2+B, and Me2+C, for the ternary ground state, 1 mM Mg2+ 300 s, and 10 mM Mn2+ 600 s structure following the previous protocol34. After there were no significant Fo-Fc peaks and each atom’s B value had roughly similar values to its ligand, we assigned occupancies for the same regions for the timepoints in between. Source data of the electron densities in r.m.s. density are provided as a Source Data file. Each structure was refined to the highest resolution data collected, which ranged between 1.47 and 1.83 Å. Software applications used in this project were compiled and configured by SBGrid67. Source data of data collection and refinement statistics are summarized in Supplementary Table 1a–f in the Source data file. All structural figures were drawn using PyMOL (http://www.pymol.org).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The data that support this study are available from the corresponding author upon reasonable request. The coordinates, density maps, and structure factors for all the structures have been deposited in Protein Data Bank (PDB) under accession codes 7U72, 7U73, 7U74, 7U75, 7U76, 7U77, 7U78, 7U79, 7U7A, 7U7B, 7U7C, 7U7D, 7U7E, 7U7F, 7U7G, 7U7I, 7U7J, 7U7K, 7U7L, 7U7R, 7U7S, 7U7T, 7U7U, 7U7V, 7U7W, 7U7X, 7U7Y, 7U7Z, 7U80, 7U81, 7U82, 7U83, and 7U84. Source data are provided with this paper.
References
Yang, W. & Gao, Y. Translesion and repair DNA polymerases: diverse structure and mechanism. Annu. Rev. Biochem. 87, 239–261 (2018).
Steitz, T. A. & Steitz, J. A. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl Acad. Sci. USA 90, 6498–6502 (1993).
Yang, W., Weng, P. J. & Gao, Y. A new paradigm of DNA synthesis: three-metal-ion catalysis. Cell Biosci. 6, 51 (2016).
Nakamura, T., Zhao, Y., Yamagata, Y., Hua, Y.-J. & Yang, W. Watching DNA polymerase η make a phosphodiester bond. Nature 487, 196–201 (2012).
Freudenthal Bret, D., Beard William, A., Shock David, D. & Wilson Samuel, H. Observing a DNA polymerase choose right from wrong. Cell 154, 157–168 (2013).
Vyas, R., Reed, A. J., Tokarsky, E. J. & Suo, Z. Viewing human DNA polymerase β faithfully and unfaithfully bypass an oxidative lesion by time-dependent crystallography. J. Am. Chem. Soc. 137, 5225–5230 (2015).
Samara, N. L. & Yang, W. Cation trafficking propels RNA hydrolysis. Nat. Struct. Mol. Biol. 25, 715–721 (2018).
Wu, J., Samara, N. L., Kuraoka, I. & Yang, W. Evolution of inosine-specific endonuclease V from bacterial DNase to eukaryotic RNase. Mol. Cell 76, 44–56.e43 (2019).
Choe, J.-Y., Poland, B. W., Fromm, H. J. & Honzatko, R. B. Role of a dynamic loop in cation activation and allosteric regulation of recombinant porcine fructose-1,6-bisphosphatase. Biochemistry 37, 11441–11450 (1998).
Aaron, J. A. & Christianson, D. W. Trinuclear metal clusters in catalysis by terpenoid synthases. Pure Appl. Chem. 82, 1585–1597 (2010).
Wang, W., Hellinga, H. W. & Beese, L. S. Structural evidence for the rare tautomer hypothesis of spontaneous mutagenesis. Proc. Natl Acad. Sci. USA 108, 17644–17648 (2011).
Kimsey, I. J. et al. Dynamic basis for dG•dT misincorporation via tautomerization and ionization. Nature 554, 195–201 (2018).
Xia, S., Wang, J. & Konigsberg, W. H. DNA mismatch synthesis complexes provide insights into base selectivity of a B family DNA polymerase. J. Am. Chem. Soc. 135, 193–202 (2013).
Nair, D. T., Johnson, R. E., Prakash, S., Prakash, L. & Aggarwal, A. K. Replication by human DNA polymerase-ι occurs by Hoogsteen base-pairing. Nature 430, 377–380 (2004).
Johnson, S. J. & Beese, L. S. Structures of mismatch replication errors observed in a DNA polymerase. Cell 116, 803–816 (2004).
Bebenek, K., Pedersen, L. C. & Kunkel, T. A. Replication infidelity via a mismatch with Watson–Crick geometry. Proc. Natl Acad. Sci. USA 108, 1862–1867 (2011).
Oertell, K. et al. Kinetic selection vs. free energy of DNA base pairing in control of polymerase fidelity. Proc. Natl Acad. Sci. USA 113, E2277 (2016).
Bebenek, K. & Kunkel, T. A. Functions of DNA polymerases. Adv. Protein Chem. 69, 137–165 (2004).
Patel, S. S., Wong, I. & Johnson, K. A. Pre-steady-state kinetic analysis of processive DNA replication including complete characterization of an exonuclease-deficient mutant. Biochemistry 30, 511–525 (1991).
Wong, I., Patel, S. S. & Johnson, K. A. An induced-fit kinetic mechanism for DNA replication fidelity: direct measurement by single-turnover kinetics. Biochemistry 30, 526–537 (1991).
Brautigam, C. A. & Steitz, T. A. Structural and functional insights provided by crystal structures of DNA polymerases and their substrate complexes. Curr. Opin. Struct. Biol. 8, 54–63 (1998).
Wu, E. Y. & Beese, L. S. The structure of a high fidelity DNA polymerase bound to a mismatched nucleotide reveals an “ajar” intermediate conformation in the nucleotide selection mechanism. J. Biol. Chem. 286, 19758–19767 (2011).
Dunlap, C. A. & Tsai, M.-D. Use of 2-aminopurine and tryptophan fluorescence as probes in kinetic analyses of DNA polymerase β †. Biochemistry 41, 11226–11235 (2002).
Rothwell, P. J., Mitaksov, V. & Waksman, G. Motions of the fingers subdomain of klentaq1 are fast and not rate limiting: implications for the molecular basis of fidelity in DNA polymerases. Mol. Cell 19, 345–355 (2005).
Tsai, Y.-C. & Johnson, K. A. A new paradigm for DNA polymerase specificity. Biochemistry 45, 9675–9687 (2006).
Raper, A. T., Reed, A. J. & Suo, Z. Kinetic mechanism of DNA polymerases: contributions of conformational dynamics and a third divalent metal ion. Chem. Rev. 118, 6000–6025 (2018).
Vande Berg, B. J., Beard, W. A. & Wilson, S. H. DNA structure and aspartate 276 influence nucleotide binding to human DNA polymerase β. J. Biol. Chem. 276, 3408–3416 (2001).
Raper, A. T. & Suo, Z. Investigation of intradomain motions of a Y-family DNA polymerase during substrate binding and catalysis. Biochemistry 55, 5832–5844 (2016).
Johnson, K. A. The kinetic and chemical mechanism of high-fidelity DNA polymerases. Biochimica et. Biophysica Acta (BBA) - Proteins Proteom. 1804, 1041–1048 (2010).
Showalter, A. K. & Tsai, M.-D. A Reexamination of the nucleotide incorporation fidelity of DNA polymerases. Biochemistry 41, 10571–10576 (2002).
Oertell, K. et al. Kinetic selection vs. free energy of DNA base pairing in control of polymerase fidelity. Proc. Natl Acad. Sci. USA 113, E2277–E2285 (2016).
Towle-Weicksel, J. B. et al. Fluorescence resonance energy transfer studies of DNA polymerase β: the critical role of fingers domain movements and a novel non-covalent step during nucleotide selection. J. Biol. Chem. 289, 16541–16550 (2014).
Alnajjar, K. S. et al. A pre-catalytic non-covalent step governs DNA polymerase β fidelity. Nucleic Acids Res. 47, 11839–11849 (2019).
Gao, Y. & Yang, W. Capture of a third Mg2+ is essential for catalyzing DNA synthesis. Science 352, 1334–1337 (2016).
Chim, N., Meza, R. A., Trinh, A. M., Yang, K. & Chaput, J. C. Following replicative DNA synthesis by time-resolved X-ray crystallography. Nat. Commun. 12, 2641 (2021).
Gregory, M. T., Gao, Y., Cui, Q. & Yang, W. Multiple deprotonation paths of the nucleophile 3′-OH in the DNA synthesis reaction. Proc. Natl Acad. Sci. USA 118, e2103990118 (2021).
Masutani, C., Kusumoto, R., Iwai, S. & Hanaoka, F. Mechanisms of accurate translesion synthesis by human DNA polymerase eta. EMBO J. 19, 3100–3109 (2000).
Limoli, C. L., Giedzinski, E., Morgan, W. F. & Cleaver, J. E. Polymerase η deficiency in the xeroderma pigmentosum variant uncovers an overlap between the S phase checkpoint and double-strand break repair. Proc. Natl Acad. Sci. USA 97, 7939 (2000).
Masutani, C. et al. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature 399, 700–704 (1999).
Barnes, R. P., Tsao, W.-C., Moldovan, G.-L. & Eckert, K. A. DNA Polymerase eta prevents tumor cell-cycle arrest and cell death during recovery from replication stress. Cancer Res. 78, 6549–6560 (2018).
Faili, A. et al. DNA polymerase η is involved in hypermutation occurring during immunoglobulin class switch recombination. J. Exp. Med. 199, 265–270 (2004).
Srivastava, A. K. et al. Enhanced expression of DNA polymerase eta contributes to cisplatin resistance of ovarian cancer stem cells. Proc. Natl Acad. Sci. USA 112, 4411–4416 (2015).
Wilson, T. M. et al. MSH2–MSH6 stimulates DNA polymerase η, suggesting a role for A:T mutations in antibody genes. J. Exp. Med. 201, 637–645 (2005).
Zeng, X., Negrete, G. A., Kasmer, C., Yang, W. W. & Gearhart, P. J. Absence of DNA polymerase η reveals targeting of C mutations on the nontranscribed strand in immunoglobulin switch regions. J. Exp. Med. 199, 917–924 (2004).
Zeng, X. et al. DNA polymerase η is an A-T mutator in somatic hypermutation of immunoglobulin variable genes. Nat. Immunol. 2, 537–541 (2001).
Yang, W., Weng, P. J. & Gao, Y. Erratum to: A new paradigm of DNA synthesis: three-metal-ion catalysis. Cell Biosci. 7, 32 (2017).
Samara, N. L., Gao, Y., Wu, J. & Yang, W. Detection of reaction intermediates in Mg(2+)-dependent DNA synthesis and RNA degradation by time-resolved X-ray crystallography. Methods Enzymol. 592, 283–327 (2017).
Zhao, Y. et al. Mechanism of somatic hypermutation at the WA motif by human DNA polymerase η. Proc. Natl Acad. Sci. USA 110, 8146 (2013).
Samara, N. L., Gao, Y., Wu, J. & Yang, W. Detection of reaction intermediates in Mg(2+)-dependent DNA synthesis and RNA degradation by time-resolved X-ray crystallography. Methods Enzymol. 592, 283–327 (2017).
Cowan, J. A. Structural and catalytic chemistry of magnesium-dependent enzymes. Biometals 15, 225–235 (2002).
Kiefer, J. R., Mao, C., Braman, J. C. & Beese, L. S. Visualizing DNA replication in a catalytically active Bacillus DNA polymerase crystal. Nature 391, 304–307 (1998).
Batra, V. K., Beard, W. A., Shock, D. D., Pedersen, L. C. & Wilson, S. H. Structures of DNA polymerase beta with active-site mismatches suggest a transient abasic site intermediate during misincorporation. Mol. Cell 30, 315–324 (2008).
Yoon, H. & Warshel, A. Simulating the fidelity and the three Mg mechanism of pol η and clarifying the validity of transition state theory in enzyme catalysis. Proteins 85, 1446–1453 (2017).
Stevens, D. R. & Hammes-Schiffer, S. Exploring the role of the third active site metal ion in DNA Polymerase η with QM/MM free energy simulations. J. Am. Chem. Soc. 140, 8965–8969 (2018).
Perera, L. et al. Requirement for transient metal ions revealed through computational analysis for DNA polymerase going in reverse. Proc. Natl Acad. Sci. USA 112, E5228–E5236 (2015).
Roston, D., Demapan, D. & Cui, Q. Extensive free-energy simulations identify water as the base in nucleotide addition by DNA polymerase. Proc. Natl Acad. Sci. USA 116, 25048–25056 (2019).
Freudenthal, B. D. et al. Uncovering the polymerase-induced cytotoxicity of an oxidized nucleotide. Nature 517, 635–639 (2015).
Reed, A. J. & Suo, Z. Time-dependent extension from an 8-oxoguanine lesion by human DNA polymerase beta. J. Am. Chem. Soc. 139, 9684–9690 (2017).
Jamsen, J. A. et al. Time-lapse crystallography snapshots of a double-strand break repair polymerase in action. Nat. Commun. 8, 253 (2017).
Donati, E., Genna, V., De & Vivo, M. Recruiting mechanism and functional role of a third metal ion in the enzymatic activity of 5′ structure-specific nucleases. J. Am. Chem. Soc. 142, 2823–2834 (2020).
Vaisman, A., Ling, H., Woodgate, R. & Yang, W. Fidelity of Dpo4: effect of metal ions, nucleotide selection and pyrophosphorolysis. EMBO J. 24, 2957–2967 (2005).
Gregory, M. T. et al. Structural and mechanistic studies of polymerase η bypass of phenanthriplatin DNA damage. Proc. Natl Acad. Sci. USA 111, 9133 (2014).
Weng, P. J. et al. Bypassing a 8,5′-cyclo-2′-deoxyadenosine lesion by human DNA polymerase η at atomic resolution. Proc. Natl Acad. Sci. USA 115, 10660 (2018).
Kabsch, W. XDS. Acta Crystallogr. Sect. D., Biol. Crystallogr. 66, 125–132 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 (2010).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 66, 486–501 (2010).
Morin, A. et al. Collaboration gets the most out of software. eLife 2, e01456 (2013).
Acknowledgements
This work is supported by CPRIT (RR190046) and the Welch Foundation (C-2033-20200401) to Y.G., and a predoctoral fellowship from the Houston Area Molecular Biophysics Program (NIH Grant No. T32 GM008280, Program Director Dr. Theodore Wensel) to C.C. Our sincere appreciation to the members of the Gao lab, and Drs. Phillips, Nikonowicz, and Lu who serve on C.C.’s thesis committee. We thank the APS LS-CAT beam technicians and research scientists Drs. Anderson, Wawrzak, and Kondrashkina. Special thanks to Dr. Wei Yang from NIH for her support in initializing the project and Dr. Mitchell Miller for advice on data collection and processing. We thank Jamie Smith, Chuxuan Li, Joshua Chang, Wei Yang, and Mitchell Miller for the critical reading of the manuscript. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817).
Author information
Authors and Affiliations
Contributions
Y.G. conceived the project. C.C. carried out all time-resolved crystallography experiments, protein purification, crystallization, data collection and processing. C.C. and C.L.L. performed the biochemical assays. C.C. and Y.G. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chang, C., Lee Luo, C. & Gao, Y. In crystallo observation of three metal ion promoted DNA polymerase misincorporation. Nat Commun 13, 2346 (2022). https://doi.org/10.1038/s41467-022-30005-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-022-30005-3
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.