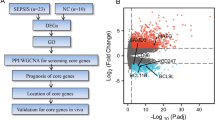
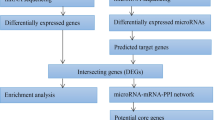
Abstract
Sepsis remains a significant global health burden and contributor to mortality, yet the precise molecular mechanisms underlying the immune response are not fully elucidated. To gain insight into this issue, we performed a comprehensive analysis using a variety of techniques including bulk RNA sequencing, single-cell RNA sequencing, and enzyme-linked immunosorbent assay (ELISA). We performed enrichment analysis of differentially expressed genes in sepsis and healthy individuals by utilizing Gene Ontology (GO) analysis and indicated significant enrichment of immune-related response. Following Weighted Gene Co-Expression Network Analysis (WGCNA) and protein-protein interaction analysis (PPI) were used to identify key immune-related hub genes and validated by ELISA to show that NLRC4 is highly expressed in sepsis. Additionally, an analysis of scRNA-seq data from newly diagnosed sepsis, sepsis diagnosis at 6 hours, and healthy samples demonstrates a significant increase in both the expression levels and proportions of NLRC4 in sepsis monocytes and neutrophils. In addition, using pySCENIC we identified upstream transcription factors that regulate NLRC4. Our study provides valuable insights into the identification of NLRC4 in peripheral blood as a potential candidate gene for the diagnosis and treatment of sepsis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 digital issues and online access to articles
$119.00 per year
only $19.83 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
No new data were created in this study and all data in the articles are annotated. All data (GSE134347, GSE65682, GSE28750, and GSE167363) and study design details in this study for the GEO can be found online at https://www.ncbi.nlm.nih.gov/geo/.
Code availability
Code available on request from corresponding author.
References
Fleischmann C, Scherag A, Adhikari NK, Hartog CS, Tsaganos T, Schlattmann P, et al. Assessment of global incidence and mortality of hospital-treated sepsis. current estimates and limitations. Am J Respiratory Crit Care Med. 2016;193:259–72.
Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3). Jama. 2016;315:801–10.
Howell MD, Davis AM. Management of sepsis and septic shock. Jama. 2017;317:847–8.
Shankar-Hari M, Phillips GS, Levy ML, Seymour CW, Liu VX, Deutschman CS, et al. Developing a new definition and assessing new clinical criteria for septic shock: for the third international consensus definitions for sepsis and septic shock (sepsis-3). Jama. 2016;315:775–87.
Gaieski DF, Edwards JM, Kallan MJ, Carr BG. Benchmarking the incidence and mortality of severe sepsis in the United States. Crit Care Med. 2013;41:1167–74.
Hershey TB, Kahn JM. State sepsis mandates - A new era for regulation of hospital quality. N. Engl J Med. 2017;376:2311–3.
Goh KH, Wang L, Yeow AYK, Poh H, Li K, Yeow JJL, et al. Artificial intelligence in sepsis early prediction and diagnosis using unstructured data in healthcare. Nat Commun. 2021;12:711.
Sinha M, Jupe J, Mack H, Coleman TP, Lawrence SM, Fraley SI. Emerging technologies for molecular diagnosis of sepsis. Clin. Microbiol. Rev. 2018;31:e00089-17.
Ji X, Li P, Fuscoe JC, Chen G, Xiao W, Shi L, et al. A comprehensive rat transcriptome built from large scale RNA-seq-based annotation. Nucleic Acids Res. 2020;48:8320–31.
Qi R, Ma A, Ma Q, Zou Q. Clustering and classification methods for single-cell RNA-sequencing data. Brief Bioinforma. 2020;21:1196–208.
Farkas JD. The complete blood count to diagnose septic shock. J Thorac Dis. 2020;12:S16–s21.
Hortová-Kohoutková M, Lázničková P, Bendíčková K, De Zuani M, Andrejčinová I, Tomášková V, et al. Differences in monocyte subsets are associated with short-term survival in patients with septic shock. J Cell Mol Med. 2020;24:12504–12.
Saviano A, Henderson NC, Baumert TF. Single-cell genomics and spatial transcriptomics: Discovery of novel cell states and cellular interactions in liver physiology and disease biology. J Hepatol. 2020;73:1219–30.
Wang Y, Cho DY, Lee H, Fear J, Oliver B, Przytycka TM. Reprogramming of regulatory network using expression uncovers sex-specific gene regulation in Drosophila. Nat Commun. 2018;9:4061.
Yang H, Cui Y, Peng W, Zhu F, Ma S, Rao M, et al. Identification of molecular subtypes and a novel prognostic model of sepsis based on ferroptosis-associated gene signature. Biomolecules. 2022;12:1479.
Scicluna BP, Uhel F, van Vught LA, Wiewel MA, Hoogendijk AJ, Baessman I, et al. The leukocyte non-coding RNA landscape in critically ill patients with sepsis. eLife. 2020;9:e58597.
Scicluna BP, Klein Klouwenberg PM, van Vught LA, Wiewel MA, Ong DS, Zwinderman AH, et al. A molecular biomarker to diagnose community-acquired pneumonia on intensive care unit admission. Am J respiratory Crit care Med. 2015;192:826–35.
Sutherland A, Thomas M, Brandon RA, Brandon RB, Lipman J, Tang B, et al. Development and validation of a novel molecular biomarker diagnostic test for the early detection of sepsis. Crit care (Lond, Engl). 2011;15:R149.
Qiu X, Li J, Bonenfant J, Jaroszewski L, Mittal A, Klein W, et al. Dynamic changes in human single-cell transcriptional signatures during fatal sepsis. J Leukoc Biol. 2021;110:1253–68.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Chen H, Boutros PC. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinforma. 2011;12:35.
Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics : a J Integr Biol. 2012;16:284–7.
Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinforma (Oxf, Engl). 2010;26:1572–3.
Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinforma (Oxf, Engl). 2016;32:2847–9.
Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinforma. 2008;9:559.
Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol Biol (Clifton, N. J). 2018;1711:243–59.
Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, et al. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902.e21.
Van de Sande B, Flerin C, Davie K, De Waegeneer M, Hulselmans G, Aibar S, et al. A scalable SCENIC workflow for single-cell gene regulatory network analysis. Nat Protoc. 2020;15:2247–76.
Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinforma. 2011;12:77.
Yuk SA, Kim H, Abutaleb NS, Dieterly AM, Taha MS, Tsifansky MD, et al. Nanocapsules modify membrane interaction of polymyxin B to enable safe systemic therapy of Gram-negative sepsis. Science Adv. 2021;7:eabj1577.
Zhang X, Cui Y, Ding X, Liu S, Han B, Duan X, et al. Analysis of mRNA‑lncRNA and mRNA‑lncRNA-pathway co‑expression networks based on WGCNA in developing pediatric sepsis. Bioengineered. 2021;12:1457–70.
Yang L, Zhou L, Li F, Chen X, Li T, Zou Z, et al. Diagnostic and prognostic value of autophagy-related key genes in sepsis and potential correlation with immune cell signatures. Front Cell Dev Biol. 2023;11:1218379.
Kofoed EM, Vance RE. NAIPs: building an innate immune barrier against bacterial pathogens. NAIPs function as sensors that initiate innate immunity by detection of bacterial proteins in the host cell cytosol. BioEssays : N. Rev Mol, Cell Dev Biol. 2012;34:589–98.
Vinzing M, Eitel J, Lippmann J, Hocke AC, Zahlten J, Slevogt H, et al. NAIP and Ipaf control Legionella pneumophila replication in human cells. J Immunol (Balt, Md : 1950). 2008;180:6808–15.
Chen Y, Wang X, Wang J, Zong J, Wan X. Revealing novel pyroptosis-related therapeutic targets for sepsis based on machine learning. BMC Med Genomics. 2023;16:23.
Cai L, Xie Y, Shao L, Hu H, Xu X, Wang H, et al. SaaS sRNA promotes Salmonella intestinal invasion via modulating MAPK inflammatory pathway. Gut microbes. 2023;15:2211184.
Lopes AH, Talbot J, Silva RL, Lima JB, França RO, Verri WA Jr, et al. Peripheral NLCR4 inflammasome participates in the genesis of acute inflammatory pain. Pain. 2015;156:451–9.
Zengini E, Hatzikotoulas K, Tachmazidou I, Steinberg J, Hartwig FP, Southam L, et al. Genome-wide analyses using UK Biobank data provide insights into the genetic architecture of osteoarthritis. Nat Genet. 2018;50:549–58.
Chabaud-Riou M, Firestein GS. Expression and activation of mitogen-activated protein kinase kinases-3 and -6 in rheumatoid arthritis. Am J Pathol. 2004;164:177–84.
Brown KA, Brain SD, Pearson JD, Edgeworth JD, Lewis SM, Treacher DF. Neutrophils in development of multiple organ failure in sepsis. Lancet (Lond, Engl). 2006;368:157–69.
Desai S, Jones SL, Turner KL, Hall J, Moore LJ. Nucleated red blood cells are associated with a higher mortality rate in patients with surgical sepsis. Surgical Infect. 2012;13:360–5.
Delano MJ, Ward PA. The immune system’s role in sepsis progression, resolution, and long-term outcome. Immunological Rev. 2016;274:330–53.
Acknowledgements
The authors gratefully acknowledge all participants and staff for their contribution to the study. The authors thank all members of the GZDlab for participating in the discussion of this artical with the first author and for their help and support.
Funding
This work is funded by China National Natural Youth Science Foundation (81802078), Zhejiang Provincial Department of Science and Technology “Leading Geese” research and development projects (2023C03083) and Zhejiang Province Public Welfare Technology Application Research Project (GF20H200021).
Author information
Authors and Affiliations
Contributions
Yinghe XU, Hongguo Zhu, and Jiaxi Chen were involved in the conception and design. Chunhui Jiang and Jiani Chen. were involved in the analysis and interpretation of the data. Jiaqing Xu, Hui Zhao, Chen Chen, and Hongguo Zhu. critically revised it for intellectual content. Chunhui Jiang, Jiani Chen, and Jiani Chen participated in drafting the manuscript. All authors agree to be accountable for all aspects of the work.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
The Ethics approval and consent to participate in the current study were obtained from the ethics committee of Taizhou Hospital of Zhejiang Province, affiliated with Wenzhou Medical University (Date: March 21, 2023/No.: K20230315), in accordance with the World Medical Association Declaration of Helsinki.
Informed consent
The data and specimens used in this study were obtained from previously collected clinical diagnostic and disease monitoring data, as well as residual samples from past patients, which were subsequently subjected to further experimental testing. The privacy and personal identity information of the subjects are protected, and there is no need for follow-up or additional information gathering from the participants. Therefore, an exemption from written informed consent was granted by the Taizhou Enze Medical Center (Group) Enze Hospital.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jiang, C., Chen, J., Xu, J. et al. Integrated analysis reveals NLRC4 as a potential biomarker in sepsis pathogenesis. Genes Immun (2024). https://doi.org/10.1038/s41435-024-00293-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41435-024-00293-4