Abstract
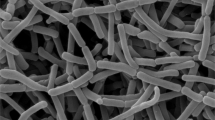
A new actinomycete strain, ODS25T, exhibited antimicrobial activity against Bacillus subtilis, Kocuria rhizophila, Staphylococcus aureus, Staphylococcus epidermidis, Candida albicans, Candida tropicalis, was isolated from the ants, Odontomachus simillimus, collected from National Science Museum Thailand, Pathum Thani, Thailand. A polyphasic technique was used to characterize the taxonomic position. The morphological and chemotaxonomic properties of the strain are typical of members of the genus Streptomyces. Strain ODS25T contained ll-diaminopimelic and glucose in the whole-cell hydrolysate. The major cellular fatty acids were iso-C16:0, iso-C15:0, and anteiso-C15:0. The polar lipids were phosphatidylethanolamine, phosphatidylinositol mannosides, phosphatidylinositol, diphosphatidylglycerol, phosphatidylglycerol, three unidentified phospholipids, three unidentified amino lipids and two unidentified lipids. The menaquinones were MK-9(H6), MK-9(H8), and MK-9(H4). The G + C content of the genomic DNA was 71.3%. The 16 S rRNA gene sequence analysis demonstrated that the strain had the highest similarity to Streptomyces lusitanus NBRC 13464T (98.07%) but shared the phylogenetic neighbour with Streptomyces sulfonofaciens JCM 5069T. Both digital DNA–DNA hybridization and average nucleotide identity values among strain ODS25T and its associated Streptomyces type strains fell within the values lower than the threshold for differentiate the strain to the same species. Based on the phenotypic characteristics and genotypic distinctiveness, strain ODS25T is considered a novel species within the genus Streptomyces, for which the name Streptomyces odontomachi sp. nov. is proposed. The type strain is ODS25T (=TBRC 16204T=NBRC 115862T).
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Parte AC, Sardà Carbasse J, Meier-Kolthoff JP, Reimer LC, Göker M. List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int J Syst Evol Microbiol. 2020;70:5607–12.
Bérdy J. Bioactive microbial metabolites. J Antibiot. 2005;58:1–26.
Abbot P. Defense in social insects: diversity, division of labor, and evolution. Annu Rev Entomol. 2022;67:407–36.
Currie CR, Scott JA, Summerbell RC, Malloch D. Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature. 1999;398:701–4.
Ye L, Zhao S, Li Y, Jiang S, Zhao Y, Li J, et al. Streptomyces lasiicapitis sp. nov., an actinomycete that produces kanchanamycin, isolated from the head of an ant (Lasius fuliginosus L.). Int J Syst Evol Microbiol. 2017;67:1529–34.
Liu C, Han C, Jiang S, Zhao X, Tian Y, Yan K, et al. Streptomyces lasii sp. nov., a novel actinomycete with antifungal activity isolated from the head of an ant (Lasius flavus). Curr Microbiol. 2018;75:353–8.
Li Y, Ye L, Wang X, Zhao J, Ma Z, Yan K, et al. Streptomyces camponoticapitis sp. nov., an actinomycete isolated from the head of an ant (Camponotus japonicus Mayr). Int J Syst Evol Microbiol. 2016;66:3855–9.
Cao T, Mu S, Lu C, Zhao S, Li D, Yan K, et al. Streptomyces amphotericinicus sp. nov., an amphotericin-producing actinomycete isolated from the head of an ant (Camponotus japonicus Mayr). Int J Syst Evol Microbiol. 2017;67:4967–73.
Tunvongvinis T, Jaitrong W, Samung Y, Tanasupawat S, Phongsopitanun W. Diversity and antimicrobial activity of the tropical ant-derived actinomycetes isolated from Thailand. AIMS Microbiol. 2024;10:68–82.
Küster E, Williams S. Selection of media for isolation of streptomycetes. Nature. 1964;202:928–9.
Hayakawa M, Nonomura H. Humic acid-vitamin agar, a new medium for the selective isolation of soil actinomycetes. J Ferment Technol. 1987;65:501–9.
Inahashi Y, Matsumoto A, Ōmura S, Takahashi Y. Streptosporangium oxazolinicum sp. nov., a novel endophytic actinomycete producing new antitrypanosomal antibiotics, spoxazomicins. J Antibiot. 2011;64:297–302.
Shirling EB, Gottlieb D. Methods for characterization of Streptomyces species. Int Syst Bacteriol. 1966;16:313–40.
Suriyachadkun C, Chunhametha S, Thawai C, Tamura T, Potacharoen W, Kirtikara K, et al. Planotetraspora thailandica sp. nov., isolated from soil in Thailand. Int J Syst Evol Microbiol. 2009;59:992–7.
Lane D 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M editors. Nucleic acid techniques in bacterial systematics. Chichester: Chichester: Wiley; 1991. p. 115-75.
Yoon S-H, Ha S-M, Kwon S, Lim J, Kim Y, Seo H, et al. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol. 2017;67:1613–7.
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol. 2018;35:1547–9.
Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–91.
Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–90.
Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13:e1005595.
Davis JJ, Wattam AR, Aziz RK, Brettin T, Butler R, Butler RM, et al. The PATRIC Bioinformatics Resource Center: expanding data and analysis capabilities. Nucleic Acids Res. 2020;48:D606–12.
Meier-Kolthoff JP, Göker M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun. 2019;10:2182.
Richter M, Rosselló-Móra R, Oliver Glöckner F, Peplies J. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics. 2016;32:929–31.
Blin K, Shaw S, Kloosterman AM, Charlop-Powers Z, Van Wezel GP, Medema MH, et al. antiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic acids Res. 2021;49:W29–35.
Kelly K Color-name charts illustrated with centroid colors. Inter-Society Color Council-National Bureau of Standards, Chicago, 1964.
Arai T, Tamotsu F, Masa H, Akihiro M, Yuzuru M Culture media for actinomycetes. Tokyo: The Society for Actinomycetes, Japan. 1975.
Williams S, Cross T Chapter XI actinomycetes. In Methods in microbiology. Vol 4. Elsevier; 1971. p. 295-334.
Gordon RE, Barnett DA, Handerhan JE, Pang CH-N. Nocardia coeliaca, Nocardia autotrophica, and the Nocardia strain. Int J Syst Bacteriol. 1974;24:54–63.
Staneck JL, Roberts GD. Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl Microbiol. 1974;28:226–31.
Collins MD, Pirouz T, Goodfellow M, Minnikin DE. Distribution of menaquinones in actinomycetes and corynebacteria. J Gen Microbiol. 1977;100:221–30.
Minnikin DE, O’donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, et al. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods. 1984;2:233–41.
Sasser M Identification of bacteria by gas chromatography of cellular fatty acids. In: MIDI technical note 101. Newark, DE: MIDI inc; 1990.
Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91.
Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA. 2009;106:19126–31.
Hu S, Li K, Zhang Y, Wang Y, Fu L, Xiao Y, et al. New insights into the threshold values of multi-locus sequence analysis, average nucleotide identity and digital DNA–DNA hybridization in delineating Streptomyces species. Front Microbiol. 2022;13:910277.
Lechevalier HA, Lechevalier MP, Gerber NN. Chemical composition as a criterion in the classification of actinomycetes. Adv Appl Microbiol. 1971;14:47–72.
Qin Z, Munnoch JT, Devine R, Holmes NA, Seipke RF, Wilkinson KA, et al. Formicamycins, antibacterial polyketides produced by Streptomyces formicae isolated from African Tetraponera plant-ants. Chem Sci. 2017;8:3218–27.
Jiang S, Piao C, Yu Y, Cao P, Li C, Yang F, et al. Streptomyces capitiformicae sp. nov., a novel actinomycete producing angucyclinone antibiotics isolated from the head of Camponotus japonicus Mayr. Int J Syst Evol Microbiol. 2018;68:118–24.
Zakalyukina YV, Osterman IA, Wolf J, Neumann-Schaal M, Nouioui I, Biryukov MV. Amycolatopsis camponoti sp. nov., new tetracenomycin-producing actinomycete isolated from carpenter ant Camponotus vagus. Antonie van Leeuwenhoek. 2022;115:533–44.
Piao C, Zheng W, Li Y, Liu C, Jin L, Song W, et al. Two new species of the genus Streptomyces: Streptomyces camponoti sp. nov. and Streptomyces cuticulae sp. nov. isolated from the cuticle of Camponotus japonicus Mayr. Arch Microbiol. 2017;199:963–70.
Liu C, Bai L, Ye L, Zhao J, Yan K, Xiang W, et al. Nocardia lasii sp. nov., a novel actinomycete isolated from the cuticle of an ant (Lasius fuliginosus L). Antonie van Leeuwenhoek. 2016;109:1513–20.
Liu C, Guan X, Li Y, Li W, Ye L, Kong X, et al. Nocardia camponoti sp. nov., an actinomycete isolated from the head of an ant (Camponotus japonicas Mayr). Int J Syst Evol Microbiol. 2016;66:1900–5.
Xiang W, Yu C, Liu C, Zhao J, Yang L, Xie B, et al. Micromonospora polyrhachis sp. nov., an actinomycete isolated from edible Chinese black ant (Polyrhachis vicina Roger). Int J Syst Evol Microbiol. 2014;64:495–500.
Liu C, Li Y, Ye L, Zhao J, Piao C, Li Z, et al. Actinocorallia lasiicapitis sp. nov., an actinomycete isolated from the head of an ant (Lasius fuliginosus L.). Int J Syst Evol Microbiol. 2016;66:2172–7.
Acknowledgements
This work was supported by the 90th Anniversary of Chulalongkorn University Scholarship under the Ratchadaphisek, Somphot Fund. We thank the 100th Anniversary Chulalongkorn University Fund, Graduated School, Chulalongkorn University for Doctoral Scholarship to T.T., and the Pharmaceutical Research Instrument Center, Faculty of Pharmaceutical Sciences, Chulalongkorn University, for providing research facilities. We thank Prof. Aharon Oren for his suggestion regarding the etymology of the type strain.
Author information
Authors and Affiliations
Contributions
TT performed screening, isolation, genotypic and phenotypic characterization, identification, and manuscript writing. WJ collected and identified the ant samples. CS and PS analyzed the chemotaxonomy. ST and WP conducted and advised all experiments.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tunvongvinis, T., Jaitrong, W., Suriyachadkun, C. et al. Streptomyces odontomachi sp. nov., a novel actinobacterium with antimicrobial potential isolated from ants (Odontomachus simillimus Smith, 1858). J Antibiot (2024). https://doi.org/10.1038/s41429-024-00766-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41429-024-00766-8