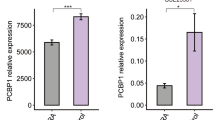
Abstract
Spliceosome dysfunction and aberrant RNA splicing underline unresolved inflammation and immunopathogenesis. Here, we revealed the misregulation of mRNA splicing via the spliceosome in the pathogenesis of rheumatoid arthritis (RA). Among them, decreased expression of RNA binding motif protein 25 (RBM25) was identified as a major pathogenic factor in RA patients and experimental arthritis mice through increased proinflammatory mediator production and increased hyperinflammation in macrophages. Multiomics analyses of macrophages from RBM25-deficient mice revealed that the transcriptional enhancement of proinflammatory genes (including Il1b, Il6, and Cxcl10) was coupled with histone 3 lysine 9 acetylation (H3K9ac) and H3K27ac modifications as well as hypoxia inducible factor-1α (HIF-1α) activity. Furthermore, RBM25 directly bound to and mediated the 14th exon skipping of ATP citrate lyase (Acly) pre-mRNA, resulting in two distinct Acly isoforms, Acly Long (Acly L) and Acly Short (Acly S). In proinflammatory macrophages, Acly L was subjected to protein lactylation on lysine 918/995, whereas Acly S did not, which influenced its affinity for metabolic substrates and subsequent metabolic activity. RBM25 deficiency overwhelmingly increased the expression of the Acly S isoform, enhancing glycolysis and acetyl-CoA production for epigenetic remodeling, macrophage overactivation and tissue inflammatory injury. Finally, macrophage-specific deletion of RBM25 led to inflammaging, including spontaneous arthritis in various joints of mice and inflammation in multiple organs, which could be relieved by pharmacological inhibition of Acly. Overall, targeting the RBM25-Acly splicing axis represents a potential strategy for modulating macrophage responses in autoimmune arthritis and aging-associated inflammation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
RNA sequencing, CUT&Tag sequencing and RIP sequencing raw data were deposited in the NCBI Gene Expression Omnibus database under the accession numbers GSE240157, GSE240158, GSE272304, GSE240159, and GSE240188, respectively.
References
Tardito S, Martinelli G, Soldano S, Paolino S, Pacini G, Patane M, et al. Macrophage M1/M2 polarization and rheumatoid arthritis: A systematic review. Autoimmun Rev. 2019;18:102397.
Udalova IA, Mantovani A, Feldmann M. Macrophage heterogeneity in the context of rheumatoid arthritis. Nat Rev Rheumatol. 2016;12:472–85.
Zhang F, Wei K, Slowikowski K, Fonseka CY, Rao DA, Kelly S, et al. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat Immunol. 2019;20:928–42.
Abramson SB, Amin A. Blocking the effects of IL-1 in rheumatoid arthritis protects bone and cartilage. Rheumatology. 2002;41:972–80.
Strand V, Kavanaugh AF. The role of interleukin-1 in bone resorption in rheumatoid arthritis. Rheumatology. 2004;43:iii10–iii16.
Liu J, Cao X. RBP-RNA interactions in the control of autoimmunity and autoinflammation. Cell Res. 2023;33:97–115.
Wu H, Gonzalez Villalobos R, Yao X, Reilly D, Chen T, Rankin M, et al. Mapping the single-cell transcriptomic response of murine diabetic kidney disease to therapies. Cell Metab. 2022;34:1064–78.e6.
Hasler R, Kerick M, Mah N, Hultschig C, Richter G, Bretz F, et al. Alterations of pre-mRNA splicing in human inflammatory bowel disease. Eur J Cell Biol. 2011;90:603–11.
Tannahill GM, Curtis AM, Adamik J, Palsson-McDermott EM, McGettrick AF, Goel G, et al. Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature. 2013;496:238–42.
Palsson-McDermott EM, Curtis AM, Goel G, Lauterbach MA, Sheedy FJ, Gleeson LE, et al. Pyruvate kinase M2 regulates Hif-1alpha activity and IL-1beta induction and is a critical determinant of the warburg effect in LPS-activated macrophages. Cell Metab. 2015;21:65–80.
Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumor growth. Nature. 2008;452:230–3.
Israelsen WJ, Dayton TL, Davidson SM, Fiske BP, Hosios AM, Bellinger G, et al. PKM2 isoform-specific deletion reveals a differential requirement for pyruvate kinase in tumor cells. Cell. 2013;155:397–409.
Ester C, Uetz P. The FF domains of yeast U1 snRNP protein Prp40 mediate interactions with Luc7 and Snu71. BMC Biochem. 2008;9:29.
Bishof I, Dammer EB, Duong DM, Kundinger SR, Gearing M, Lah JJ, et al. RNA-binding proteins with basic-acidic dipeptide (BAD) domains self-assemble and aggregate in Alzheimer’s disease. J Biol Chem. 2018;293:11047–66.
Zanoni P, Panteloglou G, Othman A, Haas JT, Meier R, Rimbert A, et al. Posttranscriptional Regulation of the Human LDL Receptor by the U2-Spliceosome. Circ Res. 2022;130:80–95.
Bramlett C, Eerdeng J, Jiang D, Lee Y, Garcia I, Vergel-Rodriguez M, et al. RNA splicing factor Rbm25 underlies heterogeneous preleukemic clonal expansion in mice. Blood. 2023;141:2961–72.
Woetzel D, Huber R, Kupfer P, Pohlers D, Pfaff M, Driesch D, et al. Identification of rheumatoid arthritis and osteoarthritis patients by transcriptome-based rule set generation. Arthritis Res Ther. 2014;16:R84.
Garcia S, Forteza J, Lopez-Otin C, Gomez-Reino JJ, Gonzalez A, Conde C. Matrix metalloproteinase-8 deficiency increases joint inflammation and bone erosion in the K/BxN serum-transfer arthritis model. Arthritis Res Ther. 2010;12:R224.
Yan M, Komatsu N, Muro R, Huynh NC, Tomofuji Y, Okada Y, et al. ETS1 governs pathological tissue-remodeling programs in disease-associated fibroblasts. Nat Immunol. 2022;23:1330–41.
Cuartero S, Weiss FD, Dharmalingam G, Guo Y, Ing-Simmons E, Masella S, et al. Control of inducible gene expression links cohesin to hematopoietic progenitor self-renewal and differentiation. Nat Immunol. 2018;19:932–41.
Soldi M, Mari T, Nicosia L, Musiani D, Sigismondo G, Cuomo A, et al. Chromatin proteomics reveals novel combinatorial histone modification signatures that mark distinct subpopulations of macrophage enhancers. Nucleic Acids Res. 2017;45:12195–213.
Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–23.
Brand DD, Latham KA, Rosloniec EF. Collagen-induced arthritis. Nat Protoc. 2007;2:1269–75.
Watson WC, Townes AS. Genetic susceptibility to murine collagen II autoimmune arthritis. Proposed relationship to the IgG2 autoantibody subclass response, complement C5, major histocompatibility complex (MHC) and non-MHC loci. J Exp Med. 1985;162:1878–91.
Zhang B, Zhang Y, Xiong L, Li Y, Zhang Y, Zhao J, et al. CD127 imprints functional heterogeneity to diversify monocyte responses in inflammatory diseases. J Exp Med. 2022;219:e20211191.
Mangan MSJ, Olhava EJ, Roush WR, Seidel HM, Glick GD, Latz E. Targeting the NLRP3 inflammasome in inflammatory diseases. Nat Rev Drug Discov. 2018;17:588–606.
Martinon F, Petrilli V, Mayor A, Tardivel A, Tschopp J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature. 2006;440:237–41.
Guo H, Ting JP. Inflammasome Assays In Vitro and in Mouse Models. Curr Protoc Immunol. 2020;131:e107.
Xiao R, Chen JY, Liang Z, Luo D, Chen G, Lu ZJ, et al. Pervasive chromatin-RNA binding protein interactions enable RNA-based regulation of transcription. Cell. 2019;178:107–21.e18.
Lin J, He Y, Chen J, Zeng Z, Yang B, Ou Q. A critical role of transcription factor YY1 in rheumatoid arthritis by regulation of interleukin-6. J Autoimmun. 2017;77:67–75.
Russell DG, Huang L, VanderVen BC. Immunometabolism at the interface between macrophages and pathogens. Nat Rev Immunol. 2019;19:291–304.
O’Neill LA, Pearce EJ. Immunometabolism governs dendritic cell and macrophage function. J Exp Med. 2016;213:15–23.
Shakespear MR, Iyer A, Cheng CY, Das Gupta K, Singhal A, Fairlie DP, et al. Lysine Deacetylases and Regulated Glycolysis in Macrophages. Trends Immunol. 2018;39:473–88.
Corcoran SE, O’Neill LA. HIF1alpha and metabolic reprogramming in inflammation. J Clin Invest. 2016;126:3699–707.
Carlson SM, Soulette CM, Yang Z, Elias JE, Brooks AN, Gozani O. RBM25 is a global splicing factor promoting inclusion of alternatively spliced exons and is itself regulated by lysine mono-methylation. J Biol Chem. 2017;292:13381–90.
Zhou A, Ou AC, Cho A, Benz EJ Jr, Huang SC. Novel splicing factor RBM25 modulates Bcl-x pre-mRNA 5’ splice site selection. Mol Cell Biol. 2008;28:5924–36.
Di Gioia M, Spreafico R, Springstead JR, Mendelson MM, Joehanes R, Levy D, et al. Endogenous oxidized phospholipids reprogram cellular metabolism and boost hyperinflammation. Nat Immunol. 2020;21:42–53.
Lauterbach MA, Hanke JE, Serefidou M, Mangan MSJ, Kolbe CC, Hess T, et al. Toll-like Receptor Signaling Rewires Macrophage Metabolism and Promotes Histone Acetylation via ATP-Citrate Lyase. Immunity. 2019;51:997–1011.e7.
Wan L, Yu W, Shen E, Sun W, Liu Y, Kong J, et al. SRSF6-regulated alternative splicing that promotes tumor progression offers a therapy target for colorectal cancer. Gut. 2019;68:118–29.
Kasowitz SD, Ma J, Anderson SJ, Leu NA, Xu Y, Gregory BD, et al. Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS Genet. 2018;14:e1007412.
Tong J, Wang X, Liu Y, Ren X, Wang A, Chen Z, et al. Pooled CRISPR screening identifies m(6)A as a positive regulator of macrophage activation. Sci Adv. 2021;7:eabd4742.
Zheng Q, Hou J, Zhou Y, Li Z, Cao X. The RNA helicase DDX46 inhibits innate immunity by entrapping m(6)A-demethylated antiviral transcripts in the nucleus. Nat Immunol. 2017;18:1094–103.
Pearce NJ, Yates JW, Berkhout TA, Jackson B, Tew D, Boyd H, et al. The role of ATP citrate-lyase in the metabolic regulation of plasma lipids. Hypolipidemic effects of SB-204990, a lactone prodrug of the potent ATP citrate-lyase inhibitor SB-201076. Biochem J. 1998;334:113–9.
Migita T, Narita T, Nomura K, Miyagi E, Inazuka F, Matsuura M, et al. ATP citrate lyase: activation and therapeutic implications in non-small cell lung cancer. Cancer Res. 2008;68:8547–54.
Wei J, Leit S, Kuai J, Therrien E, Rafi S, Harwood HJ Jr, et al. An allosteric mechanism for potent inhibition of human ATP-citrate lyase. Nature. 2019;568:566–70.
Bazilevsky GA, Affronti HC, Wei X, Campbell SL, Wellen KE, Marmorstein R. ATP-citrate lyase multimerization is required for coenzyme-A substrate binding and catalysis. J Biol Chem. 2019;294:7259–68.
Wang N, Wang W, Wang X, Mang G, Chen J, Yan X, et al. Histone Lactylation Boosts Reparative Gene Activation Post-Myocardial Infarction. Circ Res. 2022;131:893–908.
Fan M, Yang K, Wang X, Chen L, Gill PS, Ha T, et al. Lactate promotes endothelial-to-mesenchymal transition via Snail1 lactylation after myocardial infarction. Sci Adv. 2023;9:eadc9465.
Yan Y, Tao H, He J, Huang SY. The HDOCK server for integrated protein‒protein docking. Nat Protoc. 2020;15:1829–52.
Yan Y, Zhang D, Zhou P, Li B, Huang SY. HDOCK: a web server for protein‒protein and protein‒DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 2017;45:W365–W73.
Adasme MF, Linnemann KL, Bolz SN, Kaiser F, Salentin S, Haupt VJ, et al. PLIP 2021: expanding the scope of the protein‒ligand interaction profiler to DNA and RNA. Nucleic Acids Res. 2021;49:W530–W4.
Vizioli MG, Liu T, Miller KN, Robertson NA, Gilroy K, Lagnado AB, et al. Mitochondria-to-nucleus retrograde signaling drives formation of cytoplasmic chromatin and inflammation in senescence. Genes Dev. 2020;34:428–45.
Meyers AK, Zhu X. The NLRP3 Inflammasome: Metabolic Regulation and Contribution to Inflammaging. Cells. 2020;9:1808.
Havens MA, Hastings ML. Splice-switching antisense oligonucleotides as therapeutic drugs. Nucleic Acids Res. 2016;44:6549–63.
Bauman JA, Li SD, Yang A, Huang L, Kole R. Anti-tumor activity of splice-switching oligonucleotides. Nucleic Acids Res. 2010;38:8348–56.
Gao G, Xie A, Huang SC, Zhou A, Zhang J, Herman AM, et al. Role of RBM25/LUC7L3 in abnormal cardiac sodium channel splicing regulation in human heart failure. Circulation. 2011;124:1124–31.
Ge Y, Schuster MB, Pundhir S, Rapin N, Bagger FO, Sidiropoulos N, et al. The splicing factor RBM25 controls MYC activity in acute myeloid leukemia. Nat Commun. 2019;10:172.
Zhao B, Deng J, Ma M, Li N, Zhou J, Li X, et al. Environmentally relevant concentrations of 2,3,7,8-TCDD induced inhibition of multicellular alternative splicing and transcriptional dysregulation. Sci Total Environ. 2024;919:170892.
Kinyamu HK, Bennett BD, Bushel PR, Archer TK. Proteasome inhibition creates a chromatin landscape favorable to RNA Pol II processivity. J Biol Chem. 2020;295:1271–87.
Li J, Ahn JH, Wang GG. Understanding histone H3 lysine 36 methylation and its deregulation in disease. Cell Mol Life Sci. 2019;76:2899–916.
Duarte Lau F, Giugliano RP. Adenosine Triphosphate Citrate Lyase and Fatty Acid Synthesis Inhibition: A Narrative Review. JAMA Cardiol. 2023;8:879–87.
Ference BA, Ray KK, Catapano AL, Ference TB, Burgess S, Neff DR, et al. Mendelian Randomization Study of ACLY and Cardiovascular Disease. N. Engl J Med. 2019;380:1033–42.
Convertini P, Santarsiero A, Todisco S, Gilio M, Palazzo D, Pappalardo I, et al. ACLY as a modulator of liver cell functions and its role in Metabolic Dysfunction-Associated Steatohepatitis. J Transl Med. 2023;21:568.
Hochrein SM, Wu H, Eckstein M, Arrigoni L, Herman JS, Schumacher F, et al. The glucose transporter GLUT3 controls T helper 17 cell responses through glycolytic-epigenetic reprogramming. Cell Metab. 2022;34:516–32.e11.
Soto-Heredero G, Gomez de Las Heras MM, Gabande-Rodriguez E, Oller J, Mittelbrunn M. Glycolysis-a key player in the inflammatory response. FEBS J. 2020;287:3350–69.
Langston PK, Nambu A, Jung J, Shibata M, Aksoylar HI, Lei J, et al. Glycerol phosphate shuttle enzyme GPD2 regulates macrophage inflammatory responses. Nat Immunol. 2019;20:1186–95.
Verberk SGS, van der Zande HJP, Baardman J, de Goede KE, Harber KJ, Keuning ED, et al. Myeloid ATP Citrate Lyase Regulates Macrophage Inflammatory Responses In Vitro Without Altering Inflammatory Disease Outcomes. Front Immunol. 2021;12:669920.
Birch J, Gil J. Senescence and the SASP: many therapeutic avenues. Genes Dev. 2020;34:1565–76.
Lu Y, Sun Q, Guan Q, Zhang Z, He Q, He J, et al. The XOR-IDH3alpha axis controls macrophage polarization in hepatocellular carcinoma. J Hepatol. 2023;79:1172–84.
Liu Y, You Y, Lu Z, Yang J, Li P, Liu L, et al. N (6)-methyladenosine RNA modification-mediated cellular metabolism rewiring inhibits viral replication. Science. 2019;365:1171–6.
David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–8.
Xiong J, He J, Zhu J, Pan J, Liao W, Ye H, et al. Lactylation-driven METTL3-mediated RNA m(6)A modification promotes immunosuppression of tumor-infiltrating myeloid cells. Mol Cell. 2022;82:1660–77.e10.
Yang K, Fan M, Wang X, Xu J, Wang Y, Tu F, et al. Lactate promotes macrophage HMGB1 lactylation, acetylation, and exosomal release in polymicrobial sepsis. Cell Death Differ. 2022;29:133–46.
Chen L, Huang L, Gu Y, Cang W, Sun P, Xiang Y. Lactate-Lactylation Hands between Metabolic Reprogramming and Immunosuppression. Int J Mol Sci. 2022;23:11943.
Verschueren KHG, Blanchet C, Felix J, Dansercoer A, De Vos D, Bloch Y, et al. Structure of ATP citrate lyase and the origin of citrate synthase in the Krebs cycle. Nature. 2019;568:571–5.
Chen X, Birey F, Li MY, Revah O, Levy R, Thete MV, et al. Antisense oligonucleotide therapeutic approach for Timothy syndrome. Nature. 2024;628:818–25.
Ma WK, Voss DM, Scharner J, Costa ASH, Lin KT, Jeon HY, et al. ASO-based PKM splice-switching therapy inhibits hepatocellular carcinoma growth. Cancer Res. 2022;82:900–15.
Li JD, Taipale M, Blencowe BJ. Efficient, specific, and combinatorial control of endogenous exon splicing with dCasRx-RBM25. Mol Cell. 2024;84:2573–89.e5.
Zhang Y, Gao Y, Jiang Y, Ding Y, Chen H, Xiang Y, et al. Histone demethylase KDM5B licenses macrophage-mediated inflammatory responses by repressing Nfkbia transcription. Cell Death Differ. 2023;30:1279–92.
Xia M, Liu J, Wu X, Liu S, Li G, Han C, et al. Histone methyltransferase Ash1l suppresses interleukin-6 production and inflammatory autoimmune diseases by inducing the ubiquitin-editing enzyme A20. Immunity. 2013;39:470–81.
Liu X, Zhang P, Zhang Y, Wang Z, Xu S, Li Y, et al. Glycolipid iGb3 feedback amplifies innate immune responses via CD1d reverse signaling. Cell Res. 2019;29:42–53.
Wang D, Zhang Y, Xu X, Wu J, Peng Y, Li J, et al. YAP promotes the activation of NLRP3 inflammasome by blocking K27-linked polyubiquitination of NLRP3. Nat Commun. 2021;12:2674.
Liu X, Zhan Z, Li D, Xu L, Ma F, Zhang P, et al. Intracellular MHC class II molecules promote TLR-triggered innate immune responses by maintaining activation of the kinase Btk. Nat Immunol. 2011;12:416–24.
Huai W, Liu X, Wang C, Zhang Y, Chen X, Chen X, et al. KAT8 selectively inhibits antiviral immunity by acetylating IRF3. J Exp Med. 2019;216:772–85.
Zhou Q, Zhang Y, Wang B, Zhou W, Bi Y, Huai W, et al. KDM2B promotes IL-6 production and inflammatory responses through Brg1-mediated chromatin remodeling. Cell Mol Immunol. 2020;17:834–42.
Zhang D, Tang Z, Huang H, Zhou G, Cui C, Weng Y, et al. Metabolic regulation of gene expression by histone lactylation. Nature. 2019;574:575–80.
Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, et al. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–902.e21.
Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, et al. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138:114–28.
Lv D, Xu J, Qi M, Wang D, Xu W, Qiu L, et al. A strategy of screening and binding analysis of bioactive components from traditional Chinese medicine based on surface plasmon resonance biosensor. J Pharm Anal. 2022;12:500–8.
Funding
This work was supported by the National Key Research & Development Program of China (2023YFC2307302, 2019YFA0801502, 2023YFC2307001, 2023YFC2307002), the National Natural Science Foundation of China (82071790, 82070415, 82271797, 82341065, 82371825, 32400727, 82201955), the program of Shanghai outstanding academic leader in public health subject (GWVI-11.2-XD29), and the experimental animal program sponsored by the Science and Technology Commission of Shanghai Municipality (23141902300).
Author information
Authors and Affiliations
Contributions
Conceptualization: XL, ZZ, and YZ. Methodology: YZ, YG, YW, YJ, YX, BC, SX, and FM; investigation: YZ, YG, YW, YJ, YX, XW, ZW, YD, HC, BR, WH, and BC; visualization: YZ, YG, YJ, and YX; resources: SX, FM, and XR; funding acquisition: XL and ZZ; supervision: XL and ZZ; writing–original draft: YZ, XL, and ZZ; writing–review and editing: YZ, XL, and ZZ.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, Y., Gao, Y., Wang, Y. et al. RBM25 is required to restrain inflammation via ACLY RNA splicing-dependent metabolism rewiring. Cell Mol Immunol (2024). https://doi.org/10.1038/s41423-024-01212-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41423-024-01212-3