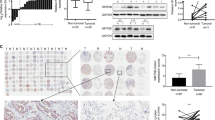
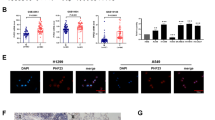
Abstract
Par6α encoded by PARD6A is a member of the PAR6 family and is reported to promote cancer initiation and progression. PARD6A is frequently upregulated in different types of cancers, but its regulatory role in lung cancer progression is yet to be established. In this study, we analyzed the PARD6A expression in biopsies from lung adenocarcinoma (LUAD) patients, and the survival probability using LUAD tissue microarray (TMA) and online datasets from TCGA and GEO. We conducted in vitro and in vivo assays to assess the role of PARD6A in regulating lung cancer progression, including proliferation, wound healing, transwell, RNA-seq, and subcutaneous tumor mice models. Our findings revealed that PARD6A is highly expressed in cancer tissues from LUAD patients and is associated with poor prognosis in LUAD patients. In vitro assays showed that PARD6A promoted cell proliferation, migration, and invasion. The transcriptome sequencing identified Serpina3 as one of the key downstream molecules of PARD6A. Ectopic expression of Serpina3 rescued impaired proliferation, migration, and invasion in PARD6A-knocking down H1299 cells, whereas silencing Serpina3 impeded enhanced proliferation, migration, and invasion in PARD6A-overexpressing H1975 cells. Our findings suggest that PARD6A promotes lung cancer progression by inducing Serpina3, which may be a promising therapeutic target.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The RNA-seq data based on the pan-Cancer Atlas run of the TCGA were obtained from the cBioPortal (https://www.cbioportal.org/). The data used for analyzing clinicopathological association are downloaded from GEO or TCGA database. The remaining data are available within the Article or Supplementary Information. Data will be made available upon request to the corresponding author.
References
Xia C, Dong X, Li H, Cao M, Sun D, He S, et al. Cancer statistics in China and United States, 2022: profiles, trends, and determinants. Chin Med J. 2022;135:584–90.
He S, Li H, Cao M, Sun D, Lei L, Li N, et al. Trends and risk factors of lung cancer in China. Chin J Cancer Res. 2020;32:683–94.
Li C, Lei S, Ding L, Xu Y, Wu X, Wang H, et al. Global burden and trends of lung cancer incidence and mortality. Chin Med J. 2023;136:1583–90.
Chan MV, Huo YR, Cao C, Ridley L. Survival outcomes for surgical resection versus CT-guided percutaneous ablation for stage I non-small cell lung cancer (NSCLC): a systematic review and meta-analysis. Eur Radiol. 2021;31:5421–33.
Halaoui R, McCaffrey L. Rewiring cell polarity signaling in cancer. Oncogene. 2015;34:939–50.
Viloria-Petit AM, David L, Jia JY, Erdemir T, Bane AL, Pinnaduwage D, et al. A role for the TGFbeta-Par6 polarity pathway in breast cancer progression. Proc Natl Acad Sci USA 2009;106:14028–33.
Zhang K, Zhao H, Ji Z, Zhang C, Zhou P, Wang L, et al. Shp2 promotes metastasis of prostate cancer by attenuating the PAR3/PAR6/aPKC polarity protein complex and enhancing epithelial-to-mesenchymal transition. Oncogene. 2016;35:1271–82.
Liu P, Zhu C, Luo J, Lan S, Su D, Wang Q, et al. Par6 regulates cell cycle progression through enhancement of Akt/PI3K/GSK-3beta signaling pathway activation in glioma. FASEB J. 2020;34:1481–96.
Togawa A, Sfakianos J, Ishibe S, Suzuki S, Fujigaki Y, Kitagawa M, et al. Hepatocyte growth factor stimulated cell scattering requires ERK and Cdc42-dependent tight junction disassembly. Biochem Biophys Res Commun. 2010;400:271–7.
Elbediwy A, Zhang Y, Cobbaut M, Riou P, Tan RS, Roberts SK, et al. The Rho family GEF FARP2 is activated by aPKCiota to control tight junction formation and polarity. J Cell Sci. 2019;132:jcs223743.
Huang Y, Liu P, Luo J, Zhu C, Lu C, Zhao N, et al. Par6 enhances glioma invasion by activating MEK/ERK pathway through a LIN28/let-7d positive feedback loop. Mol Neurobiol. 2023;60:1626–44.
Lu Z, Yuan S, Ruan L, Tu Z, Liu H. Partitioning defective 6 homolog alpha (PARD6A) promotes epithelial-mesenchymal transition via integrin beta1-ILK-SNAIL1 pathway in ovarian cancer. Cell Death Dis. 2022;13:304.
Soman A, Asha Nair S. Unfolding the cascade of SERPINA3: inflammation to cancer. Biochim Biophys Acta Rev Cancer. 2022;1877:188760.
Zhang Y, Tian J, Qu C, Peng Y, Lei J, Li K, et al. Overexpression of SERPINA3 promotes tumor invasion and migration, epithelial-mesenchymal-transition in triple-negative breast cancer cells. Breast Cancer. 2021;28:859–73.
Lara-Velazquez M, Zarco N, Carrano A, Phillipps J, Norton ES, Schiapparelli P, et al. Alpha 1-antichymotrypsin contributes to stem cell characteristics and enhances tumorigenicity of glioblastoma. Neuro Oncol. 2021;23:599–610.
Ko E, Kim JS, Bae JW, Kim J, Park SG, Jung G. SERPINA3 is a key modulator of HNRNP-K transcriptional activity against oxidative stress in HCC. Redox Biol. 2019;24:101217.
Zhou J, Zhu M, Wang Q, Deng Y, Liu N, Liu Y, et al. SERPINA3-ANKRD11-HDAC3 pathway induced aromatase inhibitor resistance in breast cancer can be reversed by HDAC3 inhibition. Commun Biol. 2023;6:695.
Santamaria M, Pardo-Saganta A, Alvarez-Asiain L, Di Scala M, Qian C, Prieto J, et al. Nuclear alpha1-antichymotrypsin promotes chromatin condensation and inhibits proliferation of human hepatocellular carcinoma cells. Gastroenterology. 2013;144:818–28.e814.
Jin Y, Zhang Y, Huang A, Chen Y, Wang J, Liu N, et al. Overexpression of SERPINA3 suppresses tumor progression by modulating SPOP/NF‑kappaB in lung cancer. Int J Oncol. 2023;63:96.
Gyorffy B. Integrated analysis of public datasets for the discovery and validation of survival-associated genes in solid tumors. Innovation. 2024;5:100625.
Gyorffy B. Transcriptome-level discovery of survival-associated biomarkers and therapy targets in non-small-cell lung cancer. Br J Pharm. 2024;181:362–74.
Szklarczyk D, Kirsch R, Koutrouli M, Nastou K, Mehryary F, Hachilif R, et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51:D638–46.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–8.
Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–5.
Lo HL, Yee JK. Production of vesicular stomatitis virus G glycoprotein (VSV-G) pseudotyped retroviral vectors. Curr Protoc Hum Genet. 2007;Chapter 12:Unit 12.7.
National Research Council. 2011. Guide for the Care and Use of Laboratory Animals: Eighth Edition. Washington, DC: The National Academies Press.
Wu H, Leng X, Liu Q, Mao T, Jiang T, Liu Y, et al. Intratumoral microbiota composition regulates chemoimmunotherapy response in esophageal squamous cell carcinoma. Cancer Res. 2023;83:3131–44.
Kemphues KJ, Priess JR, Morton DG, Cheng NS. Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell. 1988;52:311–20.
Petrie RJ, Doyle AD, Yamada KM. Random versus directionally persistent cell migration. Nat Rev Mol Cell Biol. 2009;10:538–49.
Marques E, Klefstrom J. Par6 family proteins in cancer. Oncoscience. 2015;2:894–5.
Ruan L, Shen Y, Lu Z, Shang D, Zhao Z, Lu Y, et al. Roles of partitioning-defective protein 6 (Par6) and its complexes in the proliferation, migration and invasion of cancer cells. Clin Exp Pharm Physiol. 2017;44:909–13.
Nolan ME, Aranda V, Lee S, Lakshmi B, Basu S, Allred DC, et al. The polarity protein Par6 induces cell proliferation and is overexpressed in breast cancer. Cancer Res. 2008;68:8201–9.
Aranda V, Haire T, Nolan ME, Calarco JP, Rosenberg AZ, Fawcett JP, et al. Par6-aPKC uncouples ErbB2 induced disruption of polarized epithelial organization from proliferation control. Nat Cell Biol. 2006;8:1235–45.
Karashima S, Kataoka H, Itoh H, Maruyama R, Koono M. Prognostic significance of alpha-1-antitrypsin in early stage of colorectal carcinomas. Int J Cancer. 1990;45:244–50.
Hurlimann J, van Melle G. Prognostic value of serum proteins synthesized by breast carcinoma cells. Am J Clin Pathol. 1991;95:835–43.
Luo D, Chen W, Tian Y, Li J, Xu X, Chen C, et al. Serpin peptidase inhibitor, clade A member 3 (SERPINA3), is overexpressed in glioma and associated with poor prognosis in glioma patients. Onco Targets Ther. 2017;10:2173–81.
Nimbalkar VP, Kruthika BS, Sravya P, Rao S, Sugur HS, Verma BK, et al. Differential gene expression in peritumoral brain zone of glioblastoma: role of SERPINA3 in promoting invasion, stemness and radioresistance of glioma cells and association with poor patient prognosis and recurrence. J Neurooncol. 2021;152:55–65.
Jin Y, Wang J, Ye X, Su Y, Yu G, Yang Q, et al. Identification of GlcNAcylated alpha-1-antichymotrypsin as an early biomarker in human non-small-cell lung cancer by quantitative proteomic analysis with two lectins. Br J Cancer. 2016;114:532–44.
Funamoto M, Fujio Y, Kunisada K, Negoro S, Tone E, Osugi T, et al. Signal transducer and activator of transcription 3 is required for glycoprotein 130-mediated induction of vascular endothelial growth factor in cardiac myocytes. J Biol Chem. 2000;275:10561–6.
Zhao Y, Liu Y, Zheng D. Alpha 1-antichymotrypsin/SerpinA3 is a novel target of orphan nuclear receptor Nur77. FEBS J. 2008;275:1025–38.
Yang GD, Yang XM, Lu H, Ren Y, Ma MZ, Zhu LY, et al. SERPINA3 promotes endometrial cancer cells growth by regulating G2/M cell cycle checkpoint and apoptosis. Int J Clin Exp Pathol. 2014;7:1348–58.
Li Y, Dong X, Cai J, Yin S, Sun Y, Yang D, et al. SERPINA3 induced by astroglia/microglia co-culture facilitates glioblastoma stem-like cell invasion. Oncol Lett. 2018;15:285–91.
Kulesza DW, Ramji K, Maleszewska M, Mieczkowski J, Dabrowski M, Chouaib S, et al. Search for novel STAT3-dependent genes reveals SERPINA3 as a new STAT3 target that regulates invasion of human melanoma cells. Lab Invest. 2019;99:1607–21.
Zhou ML, Chen FS, Mao H. Clinical significance and role of up-regulation of SERPINA3 expression in endometrial cancer. World J Clin Cases. 2019;7:1996–2002.
Meijers WC, Maglione M, Bakker SJL, Oberhuber R, Kieneker LM, de Jong S, et al. Heart failure stimulates tumor growth by circulating factors. Circulation. 2018;138:678–91.
Acknowledgements
This work was supported by the National Key Research and Development Program of China (Grant No. 2023YFC3402100) and the National Natural Science Foundation of China (Grant No. 92259102).
Author information
Authors and Affiliations
Contributions
LH and BT established stable knock-down and overexpressing cell lines and performed all in vitro experiments; LH and ML performed in vivo experiments; BT and DW performed bioinformatic analyses; HX and HW reconstructed knock-down and overexpression plasmids; LH, ML, BT, and XL analyzed all data and organized figures; LH, SL, and CX designed and supervised experiments; CX obtained the fundings; LH wrote the paper. All authors read and commented on the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The tumor tissue microarray (TMA) was purchased from Outdo Biotech Co., Ltd. (China). The study using the TMA was approved by the Life Sciences Ethics Committee of Outdo Biotech Co., Ltd., under the reference number: YB M-05-02. All patients and healthy donors provided informed consent to participate.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hu, L., Liu, M., Tang, B. et al. PRAD6A promotes lung adenocarcinoma cell proliferation and invasion through Serpina3. Cancer Gene Ther (2024). https://doi.org/10.1038/s41417-024-00829-w
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41417-024-00829-w