Abstract
Background
Identification of homologous recombination deficiency (HRD) remains a challenge in advanced pancreatic cancer (APC). We investigated the utility of circulating tumour DNA (ctDNA) profiling in the assessment of BRCA1/2 and ATM mutation status and treatment selection in APC.
Methods
We analysed clinical and ctDNA data of 702 patients with APC enroled in GOZILA, a ctDNA profiling study using Guardant360.
Results
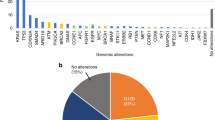
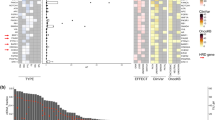
Inactivating BRCA1/2 and ATM mutations were detected in 4.8% (putative germline, 3.7%) and 4.4% (putative germline, 0.9%) of patients, respectively. Objective response (63.2% vs. 16.2%) and PFS (HR 0.55, 95% CI 0.32–0.93) on platinum-containing chemotherapy were significantly better in patients with putative germline BRCA1/2 (gBRCA) mutation than those without. In contrast, putative gBRCA mutation had no impact on the efficacy of gemcitabine plus nab-paclitaxel. In 2 patients treated with platinum-containing therapy, putative BRCA2 reversion mutations were detected. Three of seven patients with somatic BRCA mutations responded to platinum-containing therapy, while only one of four with putative germline ATM mutations did. One-third of somatic ATM mutations were in genomic loci associated with clonal haematopoiesis.
Conclusion
Comprehensive ctDNA profiling provides clinically relevant information regarding HRD status. It can be a practical, convenient option for HRD screening in APC.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
$259.00 per year
only $10.79 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
To protect the privacy and confidentiality of patients, clinical data are not made publicly available in a repository or the supplementary material of the article but will be made available following reasonable request to the corresponding author.
References
Casolino R, Paiella S, Azzolina D, Beer PA, Corbo V, Lorenzoni G, et al. Homologous recombination deficiency in pancreatic cancer: a systematic review and prevalence meta-analysis. J Clin Oncol. 2021;39:2617–31.
Lowery MA, Kelsen DP, Stadler ZK, Yu KH, Janjigian YY, Ludwig E, et al. An emerging entity: pancreatic adenocarcinoma associated with a known BRCA mutation: clinical descriptors, treatment implications, and future directions. Oncologist. 2011;16:1397–402.
Golan T, Kanji ZS, Epelbaum R, Devaud N, Dagan E, Holter S, et al. Overall survival and clinical characteristics of pancreatic cancer in BRCA mutation carriers. Br J Cancer. 2014;111:1132–8.
Rebelatto TF, Falavigna M, Pozzari M, Spada F, Cella CA, Laffi A, et al. Should platinum-based chemotherapy be preferred for germline Breast Cancer genes (BRCA) 1 and 2-mutated pancreatic ductal adenocarcinoma (PDAC) patients? A systematic review and meta-analysis. Cancer Treat Rev. 2019;80:101895.
Orsi G, Di Marco M, Cavaliere A, Niger M, Bozzarelli S, Giordano G, et al. Chemotherapy toxicity and activity in patients with pancreatic ductal adenocarcinoma and germline BRCA1-2 pathogenic variants (gBRCA1-2pv): a multicenter survey. ESMO Open. 2021;6:100238.
Golan T, Hammel P, Reni M, Van Cutsem E, Macarulla T, Hall MJ, et al. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N. Engl J Med. 2019;381:317–27.
Pishvaian MJ, Blais EM, Brody JR, Rahib L, Lyons E, De Arbeloa P, et al. Outcomes in patients with pancreatic adenocarcinoma with genetic mutations in DNA damage response pathways: results from the Know Your Tumor program. JCO Precis Oncol. 2019;3:1–10. https://doi.org/10.1200/PO.19.00115.
Park W, Chen J, Chou JF, Varghese AM, Yu KH, Wong W, et al. Genomic methods identify homologous recombination deficiency in pancreas adenocarcinoma and optimize treatment selection. Clin Cancer Res. 2020;26:3239–47.
Golan T, O’Kane GM, Denroche RE, Raitses-Gurevich M, Grant RC, Holter S, et al. Genomic features and classification of homologous recombination deficient pancreatic ductal adenocarcinoma. Gastroenterology. 2021;160:2119–32.
National Comprehensive Cancer Network. NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®): Pancreatic Adenocarcinoma. version 1.2022. February 24, 2022. https://www.nccn.org/professionals/physician_gls/pdf/pancreatic.pdf
Navina S, McGrath K, Chennat J, Singh V, Pal T, Zeh H, et al. Adequacy assessment of endoscopic ultrasound-guided, fine-needle aspirations of pancreatic masses for theranostic studies: optimization of current practices is warranted. Arch Pathol Lab Med. 2014;138:923–8.
Rothwell DG, Ayub M, Cook N, Thistlethwaite F, Carter L, Dean E, et al. Utility of ctDNA to support patient selection for early phase clinical trials: the TARGET study. Nat Med. 2019;25:738–43.
Nakamura Y, Taniguchi H, Ikeda M, Bando H, Kato K, Morizane C, et al. Clinical utility of circulating tumor DNA sequencing in advanced gastrointestinal cancer: SCRUM-Japan GI-SCREEN and GOZILA studies. Nat Med. 2020;26:1859–64.
Vidula N, Rich TA, Sartor O, Yen J, Hardin A, Nance T, et al. Routine plasma-based genotyping to comprehensively detect germline, somatic, and reversion BRCA mutations among patients with advanced solid tumors. Clin Cancer Res. 2020;26:2546–55.
Slavin TP, Banks KC, Chudova D, Oxnard GR, Odegaard JI, Nagy RJ, et al. Identification of incidental germline mutations in patients with advanced solid tumors who underwent cell-free circulating tumor DNA sequencing. J Clin Oncol. 2018;36:JCO1800328.
Hu Y, Alden RS, Odegaard JI, Fairclough SR, Chen R, Heng J, et al. Discrimination of germline EGFR T790M mutations in plasma cell-free DNA allows study of prevalence across 31,414 cancer patients. Clin Cancer Res. 2017;23:7351–9.
Nakamura Y, Fujisawa T, Taniguchi H, Bando H, Okamoto W, Tsuchihara K, et al. SCRUM-Japan GI-SCREEN and MONSTAR-SCREEN: path to the realization of biomarker-guided precision oncology in advanced solid tumors. Cancer Sci. 2021;112:4425–32.
Umemoto K, Sunakawa Y, Ueno M, Furukawa M, Mizuno N, Sudo K, et al. Clinical significance of circulating-tumour DNA analysis by metastatic sites in pancreatic cancer. Br J Cancer. 2023;128:1603–8.
Okusaka T, Nakamura M, Yoshida M, Kitano M, Ito Y, Mizuno N, et al. Clinical Practice Guidelines for Pancreatic Cancer 2022 from the Japan Pancreas Society: a synopsis. Int J Clin Oncol. 2023;28:493–511.
Kanda Y. Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transpl. 2013;48:452–8.
Momozawa Y, Sasai R, Usui Y, Shiraishi K, Iwasaki Y, Taniyama Y, et al. Expansion of cancer risk profile for BRCA1 and BRCA2 pathogenic variants. JAMA Oncol. 2022;8:871–78.
Bolton KL, Ptashkin RN, Gao T, Braunstein L, Devlin SM, Kelly D, et al. Cancer therapy shapes the fitness landscape of clonal hematopoiesis. Nat Genet. 2020;52:1219–26.
Slavin TP, Coffee B, Bernhisel R, Logan J, Cox HC, Marcucci G, et al. Prevalence and characteristics of likely-somatic variants in cancer susceptibility genes among individuals who had hereditary pan-cancer panel testing. Cancer Genet. 2019;235–236:31–8.
Xu E, Su K, Zhou Y, Gong L, Xuan Y, Liao M, et al. Comprehensive landscape and interference of clonal haematopoiesis mutations for liquid biopsy: a Chinese pan-cancer cohort. J Cell Mol Med. 2021;25:10279–90.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1.
Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019;47:D941–D947.
Stossel C, Raitses-Gurevich M, Atias D, Beller T, Glick Gorman Y, Halperin S, et al. Spectrum of response to platinum and PARP inhibitors in germline BRCA-Associated pancreatic cancer in the clinical and preclinical setting. Cancer Discov. 2023;13:1826–43.
Abrams TA, Meyer G, Meyerhardt JA, Wolpin BM, Schrag D, Fuchs CS. Patterns of chemotherapy use in a U.S.-based cohort of patients with metastatic pancreatic cancer. Oncologist. 2017;22:925–33.
Suzuki T, Kawai S, Ueno M, Lin Y, Kikuchi S. Treatment patterns in pancreatic cancer patients based on a hospital claims database in Japan. Jpn J Clin Oncol. 2021;51:228–34.
Jonsson P, Bandlamudi C, Cheng ML, Srinivasan P, Chavan SS, Friedman ND, et al. Tumour lineage shapes BRCA-mediated phenotypes. Nature. 2019;571:576–9.
Konstantinopoulos PA, Norquist B, Lacchetti C, Armstrong D, Grisham RN, Goodfellow PJ, et al. Germline and somatic tumor testing in epithelial ovarian cancer: ASCO guideline. J Clin Oncol. 2020;38:1222–45.
Abida W, Patnaik A, Campbell D, Shapiro J, Bryce AH, McDermott R, et al. Rucaparib in men with metastatic castration-resistant prostate cancer harboring a BRCA1 or BRCA2 gene alteration. J Clin Oncol. 2020;38:3763–72.
Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501.
Shroff RT, Hendifar A, McWilliams RR, Geva R, Epelbaum R, Rolfe L, et al. Rucaparib monotherapy in patients with pancreatic cancer and a known deleterious BRCA mutation. JCO Precis Oncol. 2018;2:PO.17.00316. https://doi.org/10.1200/PO.17.00316.
Lin KK, Harrell MI, Oza AM, Oaknin A, Ray-Coquard I, Tinker AV, et al. BRCA reversion mutations in circulating tumor DNA predict primary and acquired resistance to the PARP inhibitor rucaparib in high-grade ovarian carcinoma. Cancer Discov. 2019;9:210–9.
Pettitt SJ, Frankum JR, Punta M, Lise S, Alexander J, Chen Y, et al. Clinical BRCA1/2 reversion analysis identifies hotspot mutations and predicted neoantigens associated with therapy resistance. Cancer Discov. 2020;10:1475–88.
Brown TJ, Yablonovitch A, Till JE, Yen J, Kiedrowski LA, Hood R, et al. The clinical implications of reversions in patients with advanced pancreatic cancer and pathogenic variants in BRCA1, BRCA2, or PALB2 after progression on Rucaparib. Clin Cancer Res. 2023;29:5207–16.
Jensen K, Konnick EQ, Schweizer MT, Sokolova AO, Grivas P, Cheng HH, et al. Association of clonal hematopoiesis in DNA repair genes with prostate cancer plasma cell-free DNA testing interference. JAMA Oncol. 2021;7:107–10.
Acknowledgements
We would like to thank all of the patients and their families participated in this study; the Translational Research Support Section of the National Cancer Center Hospital East; and all GOZILA investigators.
Funding
This work was supported by SCRUM-Japan Funds (http://www.scrum-japan.ncc.go.jp/index.html).
Author information
Authors and Affiliations
Contributions
KS contributed to the planning and conducting of the study, recruiting patients, acquisition of data, analysis and interpretation of the data and writing of the manuscript. YN contributed to the planning and conducting of the study, analysis and interpretation of the data and writing of the manuscript. MU, MI, and CM contributed to the planning and conducting of the study, recruiting patients, acquisition of data, interpretation of the data and the revision of the manuscript. MF, N. Mizuno, YK, NO, KU, AA, MO, KO, SS, N. Matsuhashi, SI, TM, TS, HO, MG, HH, and YY, contributed to the recruitment of patients, acquisition of data and the revision of the manuscript. HB and TY contributed to the planning, conducting of the study and the revision of the manuscript. JIO contributed to analysis and interpretation of the data and the revision of the manuscript. All authors agree to be accountable for all aspects of the work and will ensure that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Corresponding author
Ethics declarations
Competing interests
KS reports honoraria from Yakult Honsha, Novartis Pharmaceuticals and Ono. Pharmaceutical; and grants for their institution from Bristol-Myers Squibb/Ono Pharmaceutical, Merck, Eisai and Incyte. YN reports advisory role from Guardant Health Pte Ltd, Natera, Inc., Roche Ltd., Seagen, Inc., Premo Partners, Inc., Daiichi Sankyo Co., Ltd., Takeda Pharmaceutical Co., Ltd., Exact Sciences Corporation, Gilead Sciences, Inc.; speakers’ bureau from Guardant Health Pte Ltd., MSD K.K, Eisai Co., Ltd., Zeria Pharmaceutical Co., Ltd., Miyarisan Pharmaceutical Co., Ltd., Merck Biopharma Co., Ltd., CareNet, Inc., Hisamitsu Pharmaceutical Co., Inc., Taiho Pharmaceutical Co., Ltd., Daiichi Sankyo Co., Ltd., Chugai Pharmaceutical Co., Ltd., Becton, Dickinson and Company, Guardant Health Japan Corp; research funding from Seagen, Inc., Genomedia Inc., Guardant Health AMEA, Inc., Guardant Health, Inc., Tempus Labs, Inc./Roche Diagnostics K.K., Daiichi Sankyo Co., Ltd., Chugai Pharmaceutical Co., Ltd. MU reports honoraria (personal fee) from Taiho Pharmaceutical, AstraZeneca, Yakult Honsha, MSD, Nihon Servier, Ono Pharmaceutical, Incyte, Chugai Pharmaceutical, Boehringer Ingelheim, J-pharma, Daiichi Sankyo, Eisai, Takeda Pharmaceutical and Novartis; and research funding (grant) from Taiho Pharmaceutical, AstraZeneca, MSD, Astellas Pharma, Eisai, Ono Pharmaceutical, Incyte, Chugai Pharmaceutical, DFP (Delta Fly Pharma), Daiichi Sankyo, Novartis, Boehringer Ingelheim, J-pharma and Chiome Bioscience. N. Mizuno has received grants or contracts from any entity from to his institution from Novartis, MSD, Incyte, Ono Pharmaceutical, Seagen, Dainippon Sumitomo Pharma and Boehringer Ingelheim; has received payment or honoraria for lectures, presentations, speakers bureaus, manuscript writing or educational events from Yakult Honsha, AstraZeneca, Novartis, FUJIFILM Toyama Chemical, Taiho Pharmaceutical and MSD; participated on a Data Safety Monitoring Board or Advisory Board for AstraZeneca. YK reports honoraria from Taiho Pharmaceutical, Merck, Yakult Honsha, Ono Pharmaceutical, Incyte, AstraZeneca and Lilly Japan; consulting or advisory role from Taiho Pharmaceutical; and research funding from Japan Agency for Medical Research and Development and Takeda. NO reports honoraria from Taiho Pharmaceutical, Eli Lilly Japan, Eisai, Bayer Yakuhin, Chugai Pharma, Ono Pharmaceutical, Takeda, Daiichi Sankyo, AstraZeneca, MSD and Incyte; advisory board from GlaxoSmithKline; and non-financial support from J-pharma. KU reports honoraria from Chugai Pharmaceutical, Taiho Pharmaceutical and Yakult Honsha. MO reports honoraria from Taiho, Ono, Yakult, Servier, MSD, Pfizer, Bayer and Novartis. SS reports grants from AstraZeneca, Incyte Corporation and Delta-Fly Pharma. TS reports grants and personal fees from Ono pharmaceutical, Chugai-Pharmaceutical, Elli-Lilly, Daiichi-Sankyo and Taiho-Pharmaceutical; and grants from Yakult-Honsha, MSD, Hutch-Med, Shionogi and Jansen, outside the submitted work. MG reports personal fees and non-financial support from Chugai Pharmaceutical, Taiho Pharmaceutical and Ono Pharmaceutical; and personal fees from Eli Lilly Japan K.K. and MSD K.K., outside the submitted work. YY reports honoraria for lectures from Ono Pharmaceutical, Taiho, Bristol Myers Squibb, Servier, Yakult, Takeda, Chugai, Daiichi Sankyo, Lilly, Incyte and Bayer. JIO is an employee and stockholder of Guardant Health. HB reports honoraria from Taiho Pharmaceutical, Eli Lilly Japan and Ono Pharmaceutical; and grants from Ono Pharmaceutical outside the submitted work. TY reports honoraria from Chugai Pharma, Takeda Pharma, Merck, Bayer Yakuhin, Ono Pharmaceutical and MSD K.K; consulting fee: Sumitomo Corp.; and research grant/funding: Amgen, Chugai, Daiichi Sankyo, Eisai, FALCO Biosystems, Genomedia Inc., Molecular Health, MSD, Nippon Boehringer Ingelheim, Ono, Pfizer, Roche Diagnostics, Sanofi, Sysmex and Taiho. MI reports honoraria from Abbott, AbbVie, AstraZeneca, Bayer, Guardant Health Japan, Bristol Myers Squibb, Chugai Pharmaceutical, EA Pharma, Eisai, Fujifilm, Incyte, Lilly Japan, MSD, Nippon Kayaku, Novartis, Ono Pharmaceutical, Nihon Servier, Taiho Pharmaceutical, Taisho Pharmaceutical Holdings, Takeda, Teijin Pharma and Yakult Honsha; advisory roles with AbbVie, AstraZeneca, Boehringer Ingelheim, Boston Scientific, Chugai Pharmaceutical, Eisai, Guardant Health Japan, Lilly Japan, MSD, Novartis, Ono Pharmaceutical and Nihon Servier; and research funding from AstraZeneca, Bayer, Bristol Myers Squibb, Chiome Bioscience, Chugai Pharmaceutical, Delta-Fly Pharma, Eisai, InVitae, J-Pharma, Lilly Japan, Merck, Merus NV, MSD, Nihon Servier, Novartis, Ono Pharmaceutical, Pfizer and Syneos Health. CM reports honoraria from Novartis, Yakult Honsha, Teijin Pharma, Taiho Pharmaceutical, Eisai, MSD K.K., and AstraZeneca; consulting fees with Yakult Honsha, MSD K.K., Servier, Boehringer Ingelheim and Taiho; and grants (for the institution) from Eisai, Yakult Honsha, Ono Pharmaceutical, Taiho Pharmaceutical, J-Pharma, AstraZeneca, Merck Biopharma, Daiichi Sankyo, HITACHI and Boehringer Ingelheim. MF, AA, KO, N. Matsuhashi, SI, TM, HO and HH have nothing to disclose.
Ethical approval and consent to participate
The study was conducted in accordance with the Declaration of Helsinki and the Japanese Ethical Guidelines for Medical and Health Research Involving Human Subjects. The protocol of the GOZILA study was approved by the Research Ethics Review Committee of National Cancer Center (National Cancer Center Hospital East; National Cancer Center Hospital) and by the ethics committee/institutional review board of other participating institutions (Aichi Cancer Center; National Hospital Organisation Kyushu Cancer Center; Hokkaido University Hospital; Saitama Cancer Center; Kanagawa Cancer Center; Kansai Rosai Hospital; National Hospital Organisation Shikoku Cancer Center; NHO Osaka National Hospital; University of Tsukuba Hospital; Chiba Cancer Center; Kyorin University Hospital; Kindai University Hospital; Kyushu University Hospital; St. Marianna University School of Medicine; Osaka University Hospital; Cancer Institute Hospital of Japanese Foundation for Cancer Research; Kobe City Medical Center General Hospital; Osaka Medical and Pharmaceutical University Hospital; Gifu University Hospital; Kanazawa University Hospital; Shizuoka Cancer Center; Kagawa University Hospital; Keio University Hospital; Saitama Medical University International Medical Center; Shimane Prefectural Central Hospital; Kansai Medical University Hospital; Kyoto Katsura Hospital; Osaka International Cancer Institute; Osaka General Medical Center). Eligible patients provided written informed consent.
Consent for publication
This manuscript does not contain any individual person’s data such as individual details, images or videos.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sudo, K., Nakamura, Y., Ueno, M. et al. Clinical utility of BRCA and ATM mutation status in circulating tumour DNA for treatment selection in advanced pancreatic cancer. Br J Cancer (2024). https://doi.org/10.1038/s41416-024-02834-0
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41416-024-02834-0