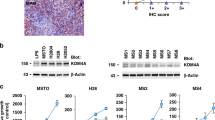
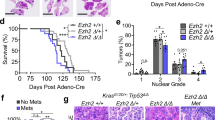
Abstract
Background
Mutational inactivation of the SETDB1 histone methyltransferase is found in a subset of mesothelioma, particularly in cases with near-haploidy and TP53 mutations. However, the tumourigenic consequences of SETDB1 inactivation are poorly understood.
Methods
In this study, we investigated SETDB1 tumour suppressor functions in mesothelioma and explored biologic relationships between SETDB1 and TP53.
Results
Immunoblotting of early passage cultures showed that SETDB1 was undetectable in 7 of 8 near-haploid mesotheliomas whereas SETDB1 expression was retained in each of 13 near-diploid mesotheliomas. TP53 aberrations were present in 5 of 8 near-haploid mesotheliomas compared to 2 of 13 near-diploid mesotheliomas, and BAP1 inactivation was demonstrated only in near-diploid mesotheliomas, indicating that near-haploid and near-diploid mesothelioma have distinct molecular and biologic profiles. Lentiviral SETDB1 restoration in near-haploid mesotheliomas (MESO257 and MESO542) reduced cell viability, colony formation, reactive oxygen species levels, proliferative marker cyclin A expression, and inhibited growth of MESO542 xenografts. The combination of SETDB1 restoration with pemetrexed and/or cisplatin treatment additively inhibited tumour growth in vitro and in vivo. Furthermore, SETDB1 restoration upregulated TP53 expression in MESO542 and MESO257, whereas SETDB1 knockdown inhibited mutant TP53 expression in JMN1B near-haploid mesothelioma cells. Likewise, TP53 knockdown inhibited SETDB1 expression. Similarly, immunoblotting evaluations of ten near-diploid mesothelioma biopsies and analysis of TCGA expression profiles showed that SETDB1 expression levels paralleled TP53 expression.
Conclusion
These findings demonstrate that SETDB1 inactivation in near-haploid mesothelioma is generally associated with complete loss of SETDB1 protein expression and dysregulates TP53 expression. Targeting SETDB1 pathways could be an effective therapeutic strategy in these often untreatable tumours.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
$259.00 per year
only $10.79 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All data generated or analysed during this study are included in this published article and its Supplementary Information files.
References
Baumann F, Ambrosi J-P, Carbone M. Asbestos is not just asbestos: an unrecognised health hazard. Lancet Oncol. 2013;14:576–8.
Craighead J. Current pathogenetic concepts of diffuse malignant mesothelioma. Hum Pathol. 1987;18:544–57.
Järvholm B, Burdorf A. Emerging evidence that the ban on asbestos use is reducing the occurrence of pleural mesothelioma in Sweden. Scand J Public Health. 2015;43:875–81.
Song W, Wang H, Lu M, Ni X, Bahri N, Zhu S, et al. AXL inactivation inhibits mesothelioma growth and migration via regulation of p53 expression. Cancers. 2020;12:2757.
Sunil SK, Prakash PN, Hariharan S, Vinod G, Preethi RT, Geetha N, et al. Adult acute lymphoblastic leukemia with near haploidy, hyperdiploidy and Ph positive lines: a rare entity with poor prognosis. Leuk Lymphoma. 2006;47:561–3.
Neragi-Miandoab S, Sugarbaker DJ. Chromosomal deletion in patients with malignant pleural mesothelioma. Interact Cardiovasc Thorac Surg. 2009;9:42–4.
Hmeljak J, Sanchez-Vega F, Hoadley KA, Shih J, Stewart C, Heiman D, et al. Integrative molecular characterization of malignant pleural mesothelioma. Cancer Discov. 2018;8:1548–65.
Sukov WR, Ketterling RP, Wei S, Monaghan K, Blunden P, Mazzara P, et al. Nearly identical near-haploid karyotype in a peritoneal mesothelioma and a retroperitoneal malignant peripheral nerve sheath tumor. Cancer Genet Cytogenet. 2010;202:123–8.
Betti M, Aspesi A, Sculco M, Matullo G, Magnani C, Dianzani I. Genetic predisposition for malignant mesothelioma: a concise review. Mutat Res Rev Mutat Res. 2019;781:1–10.
Bott M, Brevet M, Taylor BS, Shimizu S, Ito T, Wang L, et al. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat Genet. 2011;43:668–72.
Bueno R, Stawiski EW, Goldstein LD, Durinck S, De Rienzo A, Modrusan Z, et al. Comprehensive genomic analysis of malignant pleural mesothelioma identifies recurrent mutations, gene fusions and splicing alterations. Nat Genet. 2016;48:407–16.
Guo G, Chmielecki J, Goparaju C, Heguy A, Dolgalev I, Carbone M, et al. Whole-exome sequencing reveals frequent genetic alterations in BAP1, NF2, CDKN2A, and CUL1 in malignant pleural mesothelioma. Cancer Res. 2015;75:264.
Kang HC, Kim HK, Lee S, Mendez P, Kim JW, Woodard G, et al. Whole exome and targeted deep sequencing identify genome-wide allelic loss and frequent SETDB1mutations in malignant pleural mesotheliomas. Oncotarget. 2016;7:8321–31.
Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ 3rd. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–32.
Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, et al. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–31.
Minkovsky A, Sahakyan A, Rankin-Gee E, Bonora G, Patel S, Plath K. The Mbd1 - Atf7ip - Setdb1 pathway contributes to the maintenance of X chromosome inactivation. Epigenet Chromatin. 2014;7:12.
Song YJ, Choi JH, Lee H. Setdb1 is required for myogenic differentiation of C2C12 myoblast cells via maintenance of MyoD expression. Mol Cell. 2015;38:362–72.
Lawson KA, Teteak CJ, Gao J, Li N, Hacquebord J, Ghatan A, et al. ESET histone methyltransferase regulates osteoblastic differentiation of mesenchymal stem cells during postnatal bone development. FEBS Lett. 2013;587:3961–7.
Yang L, Lawson KA, Teteak CJ, Zou J, Hacquebord J, Patterson D, et al. ESET histone methyltransferase is essential to hypertrophic differentiation of growth plate chondrocytes and formation of epiphyseal plates. Dev Biol. 2013;380:99–110.
Regina C, Compagnone M, Peschiaroli A, Lena A, Annicchiarico-Petruzzelli M, Piro MC, et al. Setdb1, a novel interactor of ΔNp63, is involved in breast tumorigenesis. Oncotarget. 2016;7:28836–48.
Lafuente-Sanchis A, Zúñiga Á, Galbis JM, Cremades A, Estors M, Martínez-Hernández NJ, et al. Prognostic value of ERCC1, RRM1, BRCA1 and SETDB1 in early stage of non-small cell lung cancer. Clin Transl Oncol. 2015;18:798–804.
Sun Y, Wei M, Ren SC, Chen R, Xu WD, Wang FB, et al. Histone methyltransferase SETDB1 is required for prostate cancer cell proliferation, migration and invasion. Asian J Androl. 2014;16:319–24.
Ou WB, Corson JM, Flynn DL, Lu WP, Wise SC, Bueno R, et al. AXL regulates mesothelioma proliferation and invasiveness. Oncogene. 2011;30:1643–52.
Ou WB, Hubert C, Fletcher JA, Bueno R, Flynn DL, Sugarbaker DJ, et al. Targeted inhibition of multiple receptor tyrosine kinases in mesothelioma. Neoplasia. 2011;13:12–22.
Demetri GD, Zenzie BW, Rheinwald JG, Griffin JD. Expression of colony-stimulating factor genes by normal human mesothelial cells and human malignant mesothelioma cells lines in vitro. Blood. 1989;74:940–6.
Behbehani AM, Hunter WJ, Chapman AL, Lin F. Studies of a human mesothelioma. Hum Pathol. 1982;13:862–6.
Gordon GJ, Rockwell GN, Jensen RV, Rheinwald JG, Glickman JN, Aronson JP, et al. Identification of novel candidate oncogenes and tumor suppressors in malignant pleural mesothelioma using large-scale transcriptional profiling. Am J Pathol. 2005;166:1827–40.
Ou WB, Ni N, Zuo R, Zhuang W, Zhu M, Kyriazoglou A, et al. Cyclin D1 is a mediator of gastrointestinal stromal tumor KIT-independence. Oncogene. 2019;38:6615–29.
Chen WC, Kuang Y, Qiu HB, Cao Z, Tu Y, Sheng Q, et al. Dual targeting of insulin receptor and KIT in imatinib-resistant gastrointestinal stromal tumors. Cancer Res. 2017;77:5107–17.
Ou WB, Lu M, Eilers G, Li H, Ding J, Meng X, et al. Co-targeting of FAK and MDM2 triggers additive anti-proliferative effects in mesothelioma via a coordinated reactivation of p53. Br J Cancer. 2016;115:1253.
Zhou S, Liu L, Li H, Eilers G, Kuang Y, Shi S, et al. Multipoint targeting of the PI3K/mTOR pathway in mesothelioma. Br J Cancer. 2014;110:2479–88.
Fei Q, Shang K, Zhang J, Chuai S, Kong D, Zhou T, et al. Histone methyltransferase SETDB1 regulates liver cancer cell growth through methylation of p53. Nat Commun. 2015;6:8651.
Yoshikawa Y, Sato A, Tsujimura T, Otsuki T, Fukuoka K, Hasegawa S, et al. Biallelic germline and somatic mutations in malignant mesothelioma: multiple mutations in transcription regulators including mSWI/SNF genes. Int J Cancer. 2015;136:560–71.
Yuan L, Sun B, Xu L, Chen L, Ou W. The updating of biological functions of methyltransferase SETDB1 and its relevance in lung cancer and mesothelioma. Int J Mol Sci. 2021;22:7416.
Marcq E, Audenaerde JRV, Waele J, Jacobs J, Loenhout JV, Cavents G, et al. Building a bridge between chemotherapy and immunotherapy in malignant pleural mesothelioma: investigating the effect of chemotherapy on immune checkpoint expression. Int J Mol Sci. 2019;20:4182.
Offin M, Yang SR, Egger J, Jayakumaran G, Spencer RS, Lopardo J, et al. Molecular characterization of peritoneal mesotheliomas. J Thorac Oncol. 2022;17:455–60.
Carbone M, Adusumilli PS, Alexander HR Jr, Baas P, Bardelli F, Bononi A, et al. Mesothelioma: scientific clues for prevention, diagnosis, and therapy. CA Cancer J Clin. 2019;69:402–29.
Yoshikawa Y, Emi M, Hashimoto-Tamaoki T, Ohmuraya M, Sato A, Tsujimura T, et al. High-density array-CGH with targeted NGS unmask multiple noncontiguous minute deletions on chromosome 3p21 in mesothelioma. Proc Natl Acad Sci USA. 2016;113:13432–7.
Sun QY, Ding LW, Xiao JF, Chien W, Lim SL, Hattori N, et al. SETDB1 accelerates tumorigenesis by regulating WNT signaling pathway. J Pathol. 2015;235:559–70.
Noh HJ, Kim KA, Kim KC. p53 down-regulates SETDB1 gene expression during paclitaxel induced-cell death. Biochem Biophys Res Commun. 2014;446:43–8.
Testa JR, Cheung M, Pei J, Below JE, Tan Y, Sementino E, et al. Germline BAP1 mutations predispose to malignant mesothelioma. Nat Genet. 2011;43:1022–5.
Acknowledgements
We thank Zhejiang Provincial Key Laboratory of Silkworm Bioreactor and Biomedicine in Zhejiang Sci-Tech University for providing the experimental platform.
Funding
This research was supported by National Natural Science Foundation of China (82272695), the Key Program of Natural Science Foundation of Zhejiang Province (LZ23H160004), China. This work was also supported by the NIH/NCI SPORE 1P50CA272170-01 (JAF).
Author information
Authors and Affiliations
Contributions
WBO and JAF designed the study; MX, YT, WB, MZL, and W-BO performed the experiments and acquired the data. MX, YT, WB, MZL, JAF, and W-BO analysed and interpreted the acquired data. YT, IK, JAF, and WBO participated in scientific discussion and drafting of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Brigham and Women’s Hospital and Zhejiang Sci-Tech University Institutional Ethics Boards approved this study. Patients provided informed consent for use of their tissue samples for research purposes. The study was performed in accordance with the Declaration of Helsinki.
Consent for publication
Not applicable.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xu, M., Tu, Y., Bi, W. et al. SETDB1 tumour suppressor roles in near-haploid mesothelioma involve TP53. Br J Cancer 129, 531–540 (2023). https://doi.org/10.1038/s41416-023-02330-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-023-02330-x