Abstract
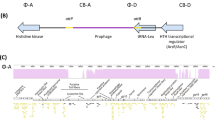
Coevolution of bacteria and phages is an important host and parasite dynamic in marine ecosystems, contributing to the understanding of bacterial community diversity. On the time scale, questions remain concerning what is the difference between phage resistance patterns in marine bacteria and how advantageous mutations gradually accumulate during coevolution. In this study, marine Aeromonas was co-cultured with its phage for 180 days and their genetic and phenotypic dynamics were measured every 30 days. We identified 11 phage resistance genes and classified them into three categories: lipopolysaccharide (LPS), outer membrane protein (OMP), and two-component system (TCS). LPS shortening and OMP mutations are two distinct modes of complete phage resistance, while TCS mutants mediate incomplete resistance by repressing the transcription of phage genes. The co-mutation of LPS and OMP was a major mode for bacterial resistance at a low cost. The mutations led to significant reductions in the growth and virulence of bacterial populations during the first 60 days of coevolution, with subsequent leveling off. Our findings reveal the marine bacterial community dynamics and evolutionary trade-offs of phage resistance during coevolution, thus granting further understanding of the interaction of marine microbes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
We are sorry, but there is no personal subscription option available for your country.
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Whole genome re-sequencing data were submitted to GenBank under BioProject ID: PRJNA972433 and PRJNA972467. The RNA-seq data were submitted to GenBank under BioProject ID: PRJNA972427. The sequence data for the A. salmonicida JNG were deposited at GenBank under accession no. CP122987-CP122989.
References
Falkowski PG, Fenchel T, Delong EF. The microbial engines that drive Earth’s biogeochemical cycles. Science. 2008;320:1034–9.
Breitbart M, Bonnain C, Malki K, Sawaya NA. Phage puppet masters of the marine microbial realm. Nat Microbiol. 2018;3:754–66.
Dennehy JJ. Bacteriophages as model organisms for virus emergence research. Trends Microbiol. 2009;17:450–7.
Hampton HG, Watson BNJ, Fineran PC. The arms race between bacteria and their phage foes. Nature. 2020;577:327–36.
Koskella B, Hernandez CA, Wheatley RM. Understanding the impacts of bacteriophage viruses: From laboratory evolution to natural ecosystems. Annu Rev Virol. 2022;9:57–8.
Markwitz P, Lood C, Olszak T, van Noort V, Lavigne R, Drulis-Kawa Z. Genome-driven elucidation of phage-host interplay and impact of phage resistance evolution on bacterial fitness. ISME J. 2021;16:533–42.
Castledine M, Padfield D, Sierocinski P, Pascual J, Hughes A, Mäkinen L, et al. Parallel evolution of Pseudomonas aeruginosa phage resistance and virulence loss in response to phage treatment in vivo and in vitro. eLife. 2022;11:e73679.
Scanlan PD, Hall AR, Blackshields G, Friman VP, Davis MR Jr., Goldberg JB, et al. Coevolution with bacteriophages drives genome-wide host evolution and constrains the acquisition of abiotic-beneficial mutations. Mol Biol Evol. 2015;32:1425–35.
Gordillo Altamirano F, Forsyth JH, Patwa R, Kostoulias X, Trim M, Subedi D, et al. Bacteriophage-resistant Acinetobacter baumannii are resensitized to antimicrobials. Nat Microbiol. 2021;6:157–61.
Bao J, Wu NN, Zeng YG, Chen LG, Li LL, Yang L, et al. Non-active antibiotic and bacteriophage synergism to successfully treat recurrent urinary tract infection caused by extensively drug-resistant Klebsiella pneumoniae. Emerg Microbes Infec. 2020;9:771–4.
Cai RP, Wang G, Lee S, Wu M, Cheng MJ, Guo ZM, et al. Three capsular polysaccharide synthesis-related glucosyltransferases, GT-1, GT-2 and WcaJ, are associated with virulence and phage sensitivity of Klebsiella pneumoniae. Front Microbiol. 2019;10:1189.
Buckling A, Wei Y, Massey RC, Brockhurst MA, Hochberg ME. Antagonistic coevolution with parasites increases the cost of host deleterious mutations. Proc Biol Sci. 2006;273:45–9.
Morgan AD, Quigley BJ, Brown SP, Buckling A. Selection on non-social traits limits the invasion of social cheats. Ecol Lett. 2012;15:841–6.
Pal C, Maciá MD, Oliver A, Schachar I, Buckling A. Coevolution with viruses drives the evolution of bacterial mutation rates. Nature. 2007;450:1079–81.
Sumrall ET, Shen Y, Keller AP, Rismondo J, Pavlou M, Eugster MR, et al. Phage resistance at the cost of virulence: Listeria monocytogenes serovar 4b requires galactosylated teichoic acids for InlB-mediated invasion. PLoS Pathog. 2019;15:e1008032.
Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8:317–27.
Tal N, Millman A, Stokar-Avihail A, Fedorenko T, Leavitt A, Melamed S, et al. Bacteria deplete deoxynucleotides to defend against bacteriophage infection. Nat Microbiol. 2022;7:1200–9.
Mumford R, Friman VP. Bacterial competition and quorum-sensing signalling shape the eco-evolutionary outcomes of model in vitro phage therapy. Evol Appl. 2017;10:161–9.
Silpe JE, Bassler BL. A host-produced quorum-sensing autoinducer controls a phage lysis-lysogeny decision. Cell. 2019;176:268–80.
Xuan G, Lin H, Tan L, Zhao G, Wang J. Quorum sensing promotes phage infection in Pseudomonas aeruginosa PAO1. mBio. 2022;13:e0317421.
Holtappels D, Alfenas-Zerbini P, Koskella B. Drivers and consequences of bacteriophage host range. Fems Microbiol Rev. 2023;47:fuad038.
de Jonge PA, Nobrega FL, Brouns SJJ, Dutilh BE. Molecular and evolutionary determinants of bacteriophage host range. Trends Microbiol. 2019;27:51–63.
Blazanin M, Turner PE. Community context matters for bacteria-phage ecology and evolution. ISME J. 2021;15:3119–28.
Xu Z, Jin P, Zhou X, Zhang Y, Wang Q, Liu X, et al. Isolation of a virulent Aeromonas salmonicida subsp. masoucida bacteriophage and its application in phage therapy in turbot (Scophthalmus maximus). Appl Environ Microbiol. 2021;87:e0146821.
Chen L, Yuan S, Liu Q, Mai G, Yang J, Deng D, et al. In vitro design and evaluation of phage cocktails against Aeromonas salmonicida. Front Microbiol. 2018;9:1476.
Kauffman KM, Polz MF. Streamlining standard bacteriophage methods for higher throughput. Methodsx. 2018;5:159–72.
Zhu P, He L, Li Y, Huang W, Xi F, Lin L, et al. OTG-snpcaller: an optimized pipeline based on TMAP and GATK for SNP calling from ion torrent data. PLoS One. 2014;9:e97507.
Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164.
Chen LH, Yang J, Yu J, Ya ZJ, Sun LL, Shen Y, et al. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 2005;33:D325–8.
Yin K, Guan Y, Ma R, Wei L, Liu B, Liu X, et al. Critical role for a promoter discriminator in RpoS control of virulence in Edwardsiella piscicida. PLoS Pathog. 2018;14:e1007272.
Kutter E. Phage host range and efficiency of plating. Methods Mol Biol. 2009;501:141–9.
Borin JM, Avrani S, Barrick JE, Petrie KL, Meyer JR. Coevolutionary phage training leads to greater bacterial suppression and delays the evolution of phage resistance. Proc Natl Acad Sci USA. 2021;118:e2104592118.
Tjaden B. De novo assembly of bacterial transcriptomes from RNA-seq data. Genome Biol. 2015;16:1.
Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–6.
Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–14.
Gong Q, Wang X, Huang H, Sun Y, Qian X, Xue F, et al. Novel host recognition mechanism of the K1 capsule-specific phage of Escherichia coli: capsular polysaccharide as the first receptor and lipopolysaccharide as the secondary receptor. J Virol. 2021;95:e0092021.
Cai RP, Wu M, Zhang H, Zhang YF, Cheng MJ, Guo ZM, et al. A smooth-type, phage-resistant Klebsiella pneumoniae mutant strain reveals that OmpC is indispensable for infection by phage GH-K3. Appl Environ Micro. 2018;84:e01585–18.
Yethon JA, Heinrichs DE, Monteiro MA, Perry MB, Whitfield C. Involvement of waaY, waaQ, and waaP in the modification of Escherichia coli lipopolysaccharide and their role in the formation of a stable outer membrane. J Biol Chem. 1998;273:26310–6.
Hameed A, Woodacre A, Machado LR, Marsden GL. An updated classification system and review of the lipooligosaccharide biosynthesis gene locus in Campylobacter jejuni. Front Microbiol. 2020;11:677.
Choudhury B, Carlson RW, Goldberg JB. The structure of the lipopolysaccharide from a galU mutant of Pseudomonas aeruginosa serogroup-O11. Carbohydr Res. 2005;340:2761–72.
Pires DP, Dötsch A, Anderson EM, Hao Y, Khursigara CM, Lam JS, et al. A genotypic analysis of five P. aeruginosa strains after biofilm infection by phages targeting different cell surface receptors. Front Microbiol. 2017;8:1229.
Gadwal S, Johnson TL, Remmer H, Sandkvist M. C-terminal processing of GlyGly-CTERM containing proteins by rhombosortase in Vibrio cholerae. PLoS Pathog. 2018;14:e1007341.
Haft DH, Varghese N. GlyGly-CTERM and rhombosortase: a C-terminal protein processing signal in a many-to-one pairing with a rhomboid family intramembrane serine protease. PLoS One. 2011;6:e28886.
Borziak K, Zhulin IB. FIST: a sensory domain for diverse signal transduction pathways in prokaryotes and ubiquitin signaling in eukaryotes. Bioinformatics. 2007;23:2518–21.
Hossain S, Boon EM. Discovery of a novel nitric oxide binding protein and nitric-oxide-responsive signaling pathway in Pseudomonas aeruginosa. ACS Infect Dis. 2017;3:454–61.
Xuan G, Lin H, Li X, Kong J, Wang J. RetS regulates phage infection in Pseudomonas aeruginosa via modulating the GacS/GacA two-component system. J Virol. 2022;96:e0019722.
Liu B, Shadrin A, Sheppard C, Mekler V, Xu Y, Severinov K, et al. A bacteriophage transcription regulator inhibits bacterial transcription initiation by σ-factor displacement. Nucleic Acids Res. 2014;42:4294–305.
Tabib-Salazar A, Mulvenna N, Severinov K, Matthews SJ, Wigneshweraraj S. Xenogeneic regulation of the bacterial transcription machinery. J Mol Biol. 2019;431:4078–92.
Woolhouse ME, Webster JP, Domingo E, Charlesworth B, Levin BR. Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nat Genet. 2002;32:569–77.
Paterson S, Vogwill T, Buckling A, Benmayor R, Spiers AJ, Thomson NR, et al. Antagonistic coevolution accelerates molecular evolution. Nature. 2010;464:275–8.
Hall AR, Scanlan PD, Morgan AD, Buckling A. Host-parasite coevolutionary arms races give way to fluctuating selection. Ecol Lett. 2011;14:635–42.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 32025038), the Shanghai Frontiers Science Center of Optogenetic Techniques for Cell Metabolism grant 2021 Sci & Tech 03-28 (Shanghai Municipal Education Commission), China Agriculture Research System of MOF and MARA (CARS-47), and ECUST-OPM Innovation Institute Open Fund (20230801).
Author information
Authors and Affiliations
Contributions
Conceptualization: XZ DZ; Data curation: XZ DZ. Funding acquisition: LQ ZY; Investigation: XZ DZ SL XY; Project administration: LQ ZY WZ; Supervision: LQ WZ; Validation: XZ DZ SL; Visualization: XZ; Writing – original draft: XZ DZ; Writing – review and editing: XZ LQ ZY.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xu, Z., Ding, Z., Shi, L. et al. Coevolution between marine Aeromonas and phages reveals temporal trade-off patterns of phage resistance and host population fitness. ISME J 17, 2200–2209 (2023). https://doi.org/10.1038/s41396-023-01529-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-023-01529-3