Abstract
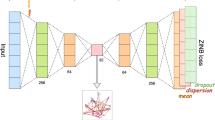
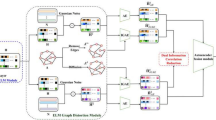
Single-cell transcriptome sequencing (scRNA-seq) is a high-throughput technique used to study gene expression at the single-cell level. Clustering analysis is a commonly used method in scRNA-seq data analysis, helping researchers identify cell types and uncover interactions between cells. However, the choice of a robust similarity metric in the clustering procedure is still an open challenge due to the complex underlying structures of the data and the inherent noise in data acquisition. Here, we propose a deep clustering method for scRNA-seq data called scRISE (scRNA-seq Iterative Smoothing and self-supervised discriminative Embedding model) to resolve this challenge. The model consists of two main modules: an iterative smoothing module based on graph autoencoders designed to denoise the data and refine the pairwise similarity in turn to gradually incorporate cell structural features and enrich the data information; and a self-supervised discriminative embedding module with adaptive similarity threshold for partitioning samples into correct clusters. Our approach has shown improved quality of data representation and clustering on seventeen scRNA-seq datasets against a number of state-of-the-art deep learning clustering methods. Furthermore, utilizing the scRISE method in biological analysis against the HNSCC dataset has unveiled 62 informative genes, highlighting their potential roles as therapeutic targets and biomarkers.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The authors declare that all data supporting the findings of this study are available within the article or from the corresponding author upon reasonable request.
Code availability
The scRISE software package and source code are available in Github (https://github.com/LiLab-ssruan/scRISE).
References
Hwang B, Lee JH, Bang D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp Mol Med. 2018;50:1–14.
Wen L, Li G, Huang T, Geng W, Pei H, Yang J, et al. Single-cell technologies: from research to application. Innovation. 2022;3:100342.
Eraslan G, Drokhlyansky E, Anand S, Fiskin E, Subramanian A, Slyper M, et al. Single-nucleus cross-tissue molecular reference maps toward understanding disease gene function. Science. 2022;376:eabl4290.
Iacono G, Mereu E, Guillaumet-Adkins A, Corominas R, Cuscó I, Rodríguez-Esteban G, et al. bigSCale: an analytical framework for big-scale single-cell data. Genome Res. 2018;28:878–90.
Chen G, Ning B, Shi T. Single-cell RNA-seq technologies and related computational data analysis. Front Genet. 2019;10:317.
Pal B, Chen Y, Vaillant F, Jamieson P, Gordon L, Rios AC, et al. Construction of developmental lineage relationships in the mouse mammary gland by single-cell RNA profiling. Nat Commun. 2017;8:1627.
Wen J, Ling R, Chen R, Zhang S, Dai Y, Zhang T, et al. Diversity of arterial cell and phenotypic heterogeneity induced by high-fat and high-cholesterol diet. Front Cell Dev Biol. 2023;11:971091.
Yang L, Liu J, Lu Q, Riggs AD, Wu X. SAIC: an iterative clustering approach for analysis of single cell RNA-seq data. BMC Genomics. 2017;18:689.
Pal S, Mondal S, Das G, Khatua S, Ghosh Z. Big data in biology: The hope and present-day challenges in it. Gene Rep. 2020;21:100869.
Lingxue Z, Jing L, Bernie D, Kathryn R. A unified statistical framework for single cell and bulk RNA sequencing data. Ann Appl Stat. 2018;12:609–32.
Kharchenko PV, Silberstein L, Scadden DT. Bayesian approach to single-cell differential expression analysis. Nat Methods. 2014;11:740–2.
Tung P-Y, Blischak JD, Hsiao CJ, Knowles DA, Burnett JE, Pritchard JK, et al. Batch effects and the effective design of single-cell gene expression studies. Sci Rep. 2017;7:39921.
žurauskienė J, Yau C. pcaReduce: hierarchical clustering of single cell transcriptional profiles. BMC Bioinform. 2016;17:140.
Lin P, Troup M, Ho JWK. CIDR: Ultrafast and accurate clustering through imputation for single-cell RNA-seq data. Genome Biol. 2017;18:59.
Wang B, Ramazzotti D, De Sano L, Zhu J, Pierson E, Batzoglou S. SIMLR: a tool for large-scale genomic analyses by multi-kernel learning. Proteomics. 2018;18:1700232.
Kiselev VY, Kirschner K, Schaub MT, Andrews T, Yiu A, Chandra T, et al. SC3: consensus clustering of single-cell RNA-seq data. Nat Methods. 2017;14:483–6.
Eraslan G, Simon LM, Mircea M, Mueller NS, Theis FJ. Single-cell RNA-seq denoising using a deep count autoencoder. Nat Commun. 2019;10:390.
Tian T, Wan J, Song Q, Wei Z. Clustering single-cell RNA-seq data with a model-based deep learning approach. Nat Mach Intell. 2019;1:191–8.
Chen L, Wang W, Zhai Y, Deng M. Deep soft K-means clustering with self-training for single-cell RNA sequence data. NAR Genomics Bioinform. 2020;2:lqaa039.
Lopez R, Regier J, Cole MB, Jordan MI, Yosef N. Deep generative modeling for single-cell transcriptomics. Nat Methods. 2018;15:1053–8.
Wang J, Ma A, Chang Y, Gong J, Jiang Y, Qi R, et al. scGNN is a novel graph neural network framework for single-cell RNA-Seq analyses. Nat Commun. 2021;12:1882.
Luo Z, Xu C, Zhang Z, Jin W. A topology-preserving dimensionality reduction method for single-cell RNA-seq data using graph autoencoder. Sci Rep. 2021;11:20028.
Yu Z, Lu Y, Wang Y, Tang F, Wong K-C, Li X. ZINB-based graph embedding autoencoder for single-cell RNA-seq interpretations. Proc AAAI Confer Artif Intell. 2022;36:4671–9.
Gan Y, Huang X, Zou G, Zhou S, Guan J. Deep structural clustering for single-cell RNA-seq data jointly through autoencoder and graph neural network. Brief Bioinform. 2022;23:bbac018.
Yu B, Chen C, Qi R, Zheng RQ, Skillman-Lawrence PJ, Wang XL, et al. scGMAI: a Gaussian mixture model for clustering single-cell RNA-Seq data based on deep autoencoder. Brief Bioinform. 2021;22:bbaa316.
Lee DD, Seung HS. Learning the parts of objects by non-negative matrix factorization. Nature. 1999;401:788–91.
Tibshirani R. Regression shrinkage and selection via the lasso. J R Stat Soc: Ser B (Methodol). 1996;58:267–88.
Yu J, Liang QY, Wang J, Cheng Y, Wang S, Poon TCW, et al. Zinc-finger protein 331, a novel putative tumor suppressor, suppresses growth and invasiveness of gastric cancer. Oncogene. 2013;32:307–17.
Wang J, Zhang G, Sui Y, Yang Z, Chu Y, Tang H, et al. CD52 is a prognostic biomarker and associated with tumor microenvironment in breast cancer. Front Genet. 2020;11:578002.
Ma Y-F, Chen Y, Fang D, Huang Q, Luo Z, Qin Q, et al. The immune-related gene CD52 is a favorable Biomark breast cancer prognosis. Gland Surg. 2021;10:780–98.
Byerly JH, Port ER, Irie HY. PRKCQ inhibition enhances chemosensitivity of triple-negative breast cancer by regulating Bim. Breast Cancer Res. 2020;22:72.
Katzendorn O, Peters I, Dubrowinskaja N, Tezval H, Tabrizi PF, von Klot CA, et al. DNA methylation of tumor associated calcium signal transducer 2 (TACSTD2) loci shows association with clinically aggressive renal cell cancers. BMC Cancer. 2021;21:444.
Chaplin DD. Overview of the immune response. J Allergy Clin Immunol. 2010;125:S3–S23.
Greenlee JD, Subramanian T, Liu K, King MR. Rafting down the metastatic cascade: the role of lipid rafts in cancer metastasis, cell death, and clinical outcomes. Cancer Res. 2021;81:5–17.
DiRusso CJ, Dashtiahangar M, Gilmore TD. Scaffold proteins as dynamic integrators of biological processes. J Biol Chem. 2022;298:102628.
Shah K, Al-Haidari A, Sun J, Kazi JU. T cell receptor (TCR) signaling in health and disease. Signal Transduct Target Ther. 2021;6:412.
Bhaumik S, Basu R. Cellular and molecular dynamics of Th17 differentiation and its developmental plasticity in the intestinal immune response. Front Immunol. 2017;8:254.
Kim MJ, Ha S-J. Differential role of PD-1 expressed by various immune and tumor cells in the tumor immune microenvironment: expression, function, therapeutic efficacy, and resistance to cancer immunotherapy. Front Cell Dev Biol. 2021;9:767466.
Zappia L, Phipson B, Oshlack A. Splatter: simulation of single-cell RNA sequencing data. Genome Biol. 2017;18:174.
Deng Q, Ramsköld D, Reinius B, Sandberg R. Single-cell RNA-Seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science. 2014;343:193–6.
Hayashi T, Ozaki H, Sasagawa Y, Umeda M, Danno H, Nikaido I. Single-cell full-length total RNA sequencing uncovers dynamics of recursive splicing and enhancer RNAs. Nat Commun. 2018;9:619.
Schaum N, Karkanias J, Neff NF, May AP, Quake SR, Wyss-Coray T, et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 2018;562:367–72.
Baron M, Veres A, Wolock Samuel L, Faust Aubrey L, Gaujoux R, Vetere A, et al. A single-cell transcriptomic map of the human and mouse pancreas reveals inter- and intra-cell population structure. Cell Syst. 2016;3:346–360.e344.
Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–201.
Romanov RA, Zeisel A, Bakker J, Girach F, Hellysaz A, Tomer R, et al. Molecular interrogation of hypothalamic organization reveals distinct dopamine neuronal subtypes. Nat Neurosci. 2017;20:176–88.
Zeisel A, Muñoz-Manchado AB, Codeluppi S, Lönnerberg P, La Manno G, Juréus A, et al. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–42.
Li H, Courtois ET, Sengupta D, Tan Y, Chen KH, Goh JJL, et al. Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat Genet. 2017;49:708–18.
Chu L-F, Leng N, Zhang J, Hou Z, Mamott D, Vereide DT, et al. Single-cell RNA-seq reveals novel regulators of human embryonic stem cell differentiation to definitive endoderm. Genome Biol. 2016;17:173.
Petropoulos S, Edsgärd D, Reinius B, Deng Q, Panula SaritaP, Codeluppi S, et al. Single-Cell RNA-seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell. 2016;165:1012–26.
Puram SV, Tirosh I, Parikh AS, Patel AP, Yizhak K, Gillespie S, et al. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell. 2017;171:1611–1624.e1624.
Tirosh I, Izar B, Prakadan SM, Wadsworth MH, Treacy D, Trombetta JJ, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–96.
Tosches MA, Yamawaki TM, Naumann RK, Jacobi AA, Tushev G, Laurent G. Evolution of pallium, hippocampus, and cortical cell types revealed by single-cell transcriptomics in reptiles. Science. 2018;360:881–8.
Bach K, Pensa S, Grzelak M, Hadfield J, Adams DJ, Marioni JC, et al. Differentiation dynamics of mammary epithelial cells revealed by single-cell RNA sequencing. Nat Commun. 2017;8:2128.
Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19:15.
Taubin G. A signal processing approach to fair surface design. In: Proceedings of the 22nd Annual Conference on Computer Graphics and Interactive Techniques. Association for Computing Machinery (ACM); 1995. p. 351–8.
Cui G, Zhou J, Yang C, Liu Z. Adaptive graph encoder for attributed graph embedding. In: Proceedings of the 26th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining. Association for Computing Machinery: Virtual Event, CA, USA, 2020, pp 976-85.
Funding
This work was supported in part by the National Natural Science Foundation of China (grants 82425104 and 82150208 to HL, 82173690 to SLL); the National Key R&D Program of China (2022YFC3400501, 2022YFC3400504); SLL is also sponsored by the Shanghai Rising-Star Program (23QA1402800).
Author information
Authors and Affiliations
Contributions
Jinxin Xie: Conceptualization, methodology, software, writing - original draft. Shanshan Ruan: Data curation, formal analysis, visualization, writing - review & editing. Mingyan Tu: Validation, investigation, software. Zhen Yuan, Jianguo Hu: Investigation. Honglin Li, Shiliang Li: Supervision, project administration, funding acquisition.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
41388_2024_3074_MOESM1_ESM.docx
Supporting Information for Clustering single-cell RNA sequencing data via iterative smoothing and self-supervised discriminative embedding
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xie, J., Ruan, S., Tu, M. et al. Clustering single-cell RNA sequencing data via iterative smoothing and self-supervised discriminative embedding. Oncogene 43, 2279–2292 (2024). https://doi.org/10.1038/s41388-024-03074-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-024-03074-5