Abstract
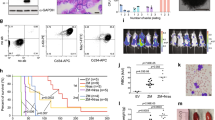
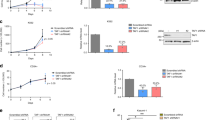
The t(8;21) fusion product, AML1/ETO, and hypoxia-inducible factor 1α (HIF1α) form a feed-forward transcription loop that cooperatively transactivates the DNA methyltransferase 3a gene promoter that leads to DNA hypermethylation and drives leukemia cell growth. Suppression of the RNA N6-methyladenosine (m6A)-reader enzyme YTH N6-methyladenosine RNA binding protein 2 (YTHDF2) specifically compromises cancer stem cells in acute myeloid leukemia (AML) but promotes hematopoietic stem cell expansion without derailing normal hematopoiesis. However, the relevance of expression between AML1/ETO-HIF1α loop and YTHDF2, and its functional relationship with t(8;21) AML have not been documented. Here, we show that YTHDF2 is highly expressed in t(8;21) AML patients and associated with a higher risk of relapse and inferior relapse-free survival. Knockdown of YTHDF2 in leukemia cells causes an impaired cell proliferation rate in vitro and in mice. Mechanistically, HIF1α is able to bind to the hypoxia-response elements of the 5′-untranslated region of the YTHDF2 gene and promotes the transactivity of the YTHDF2 promoter. Knockdown and overexpression of either AML1/ETO or HIF1α resulted in decreased and increased YTHDF2 protein and mRNA expression in t(8;21) AML cells. In particular, knockdown of YTHDF2 resulted in increased global mRNA m6A levels in t(8;21) AML cells, accompanied by increased TNF receptor superfamily member 1b (TNFRSF1b) mRNA and protein expression levels. Last, we demonstrated that the m6A methylation and expression levels of the TNFRSF1b gene were both negatively correlated with HIF1α expression levels. In conclusion, YTHDF2 is a downstream target of the AML1/ETO-HIF1α loop and promotes cell proliferation probably by modulating the global m6A methylation in t(8;21) AML.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The m6A sequencing data have been deposited in Gene Expression Omnibus (GEO) with the accession code GSE168778.
References
Appelbaum FR, Kopecky KJ, Tallman MS, Slovak ML, Gundacker HM, Kim HT, et al. The clinical spectrum of adult acute myeloid leukaemia associated with core binding factor translocations. Br J Haematol. 2006;135:165–73.
Schlenk RF, Benner A, Krauter J, Büchner T, Sauerland C, Ehninger G, et al. Individual patient data-based meta-analysis of patients aged 16 to 60 years with core binding factor acute myeloid leukemia: a survey of the German Acute Myeloid Leukemia Intergroup. J Clin Oncol. 2004;22:3741–50.
Marcucci G, Mrózek K, Ruppert AS, Maharry K, Kolitz JE, Moore JO, et al. Prognostic factors and outcome of core binding factor acute myeloid leukemia patients with t(8;21) differ from those of patients with inv(16): a Cancer and Leukemia Group B study. J Clin Oncol. 2005;23:5705–17.
Prébet T, Boissel N, Reutenauer S, Thomas X, Delaunay J, Cahn JY, et al. Acute myeloid leukemia with translocation (8;21) or inversion (16) in elderly patients treated with conventional chemotherapy: a collaborative study of the French CBF-AML intergroup. J Clin Oncol. 2009;27:4747–53.
Yan M, Burel SA, Peterson LF, Kanbe E, Iwasaki H, Boyapati A, et al. Deletion of an AML1-ETO C-terminal NcoR/SMRT-interacting region strongly induces leukemia development. Proc Natl Acad Sci USA. 2004;101:17186–91.
Al-Harbi S, Aljurf M, Mohty M, Almohareb F, Ahmed SOA. An update on the molecular pathogenesis and potential therapeutic targeting of AML with t(8;21)(q22;q22.1); RUNX1-RUNX1T1. Blood Adv. 2020;4:229–38.
Gao XN, Yan F, Lin J, Gao L, Lu XL, Wei SC, et al. AML1/ETO cooperates with HIF1α to promote leukemogenesis through DNMT3a transactivation. Leukemia. 2015;29:1730–40.
Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–99.
Barbieri I, Tzelepis K, Pandolfini L, Shi J, Millán-Zambrano G, Robson SC, et al. Promoter-bound METTL3 maintains myeloid leukaemia by m6A-dependent translation control. Nature. 2017;552:126–31.
Vu LP, Pickering BF, Cheng Y, Zaccara S, Nguyen D, Minuesa G, et al. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med. 2017;23:1369–76.
Weng H, Huang H, Wu H, Qin X, Zhao BS, Dong L, et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m6A modification. Cell Stem Cell. 2018;22:191–205.
Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, et al. FTO plays an oncogenic role in acute myeloid leukemia as a n6-methyladenosine RNA demethylase. Cancer Cell. 2017;31:127–41.
Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, et al. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016;7:12626.
Li Z, Qian P, Shao W, Shi H, He XC, Gogol M, et al. Suppression of m6A reader Ythdf2 promotes hematopoietic stem cell expansion. Cell Res. 2018;28:904–17.
Wang H, Zuo H, Liu J, Wen F, Gao Y, Zhu X, et al. Loss of YTHDF2-mediated m6A-dependent mRNA clearance facilitates hematopoietic stem cell regeneration. Cell Res. 2018;28:1035–8.
Paris J, Morgan M, Campos J, Spencer GJ, Shmakova A, Ivanova I, et al. Targeting the RNA m6A reader YTHDF2 selectively compromises cancer stem cells in acute myeloid leukemia. Cell Stem Cell. 2019;25:137–48.
de Jonge HJ, Valk PJ, Veeger NJ, ter Elst A, den Boer ML, Cloos J, et al. High VEGFC expression is associated with unique gene expression profiles and predicts adverse prognosis in pediatric and adult acute myeloid leukemia. Blood. 2010;116:1747–54.
Valk PJ, Verhaak RG, Beijen MA, Erpelinck CA, Barjesteh van Waalwijk van Doorn-Khosrovani S, Boer JM, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med. 2004;350:1617–28.
Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y, et al. R-2HG exhibits anti-tumor activity by targeting FTO/m6A/MYC/CEBPA signaling. Cell. 2018;172:90–105.
Zhong L, Liao D, Zhang M, Zeng C, Li X, Zhang R, et al. YTHDF2 suppresses cell proliferation and growth via destabilizing the EGFR mRNA in hepatocellular carcinoma. Cancer Lett. 2019;442:252–61.
Hou J, Zhang H, Liu J, Zhao Z, Wang J, Lu Z, et al. YTHDF2 reduction fuels inflammation and vascular abnormalization in hepatocellular carcinoma. Mol Cancer. 2019;18:163.
Sheng H, Li Z, Su S, Sun W, Zhang X, Li L, et al. YTH domain family 2 promotes lung cancer cell growth by facilitating 6-phosphogluconate dehydrogenase mRNA translation. Carcinogenesis. 2020;41:541–50.
Li J, Meng S, Xu M, Wang S, He L, Xu X. et al. Downregulation of N(6)-methyladenosine binding YTHDF2 protein mediated by miR-493-3p suppresses prostate cancer by elevating N(6)-methyladenosine levels. Oncotarget. 2017;9:3752–64.
Chen J, Sun Y, Xu X, Wang D, He J, Zhou H, et al. YTH domain family 2 orchestrates epithelial-mesenchymal transition/proliferation dichotomy in pancreatic cancer cells. Cell Cycle. 2017;16:2259–71.
Wang J, Hoshino T, Redner RL, Kajigaya S, Liu JM. ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex. Proc Natl Acad Sci USA. 1998;95:10860–5.
Fazi F, Racanicchi S, Zardo G, Starnes LM, Mancini M, Travaglini L, et al. Epigenetic silencing of the myelopoiesis regulator microRNA-223 by the AML1/ETO oncoprotein. Cancer Cell. 2007;12:457–66.
Wang L, Gural A, Sun XJ, Zhao X, Perna F, Huang G, et al. The leukemogenicity of AML1-ETO is dependent on site-specific lysine acetylation. Science. 2011;333:765–9.
Shia WJ, Okumura AJ, Yan M, Sarkeshik A, Lo MC, Matsuura S, et al. PRMT1 interacts with AML1-ETO to promote its transcriptional activation and progenitor cell proliferative potential. Blood. 2012;119:4953–62.
Hou J, Zhang H, Liu J, Zhao Z, Wang J, Lu Z, et al. Hypoxia regulates m6A-reader YTHDF2 to fuel cancer-promoting inflammation in hepatocellular carcinoma. Cancer Res. 2019;79(13 Suppl):Abstract nr 4319.
Kühnl A, Gökbuget N, Kaiser M, Schlee C, Stroux A, Burmeister T. et al. Overexpression of LEF1 predicts unfavorable outcome in adultpatients with B-precursor acute lymphoblastic leukemia. Blood. 2011;118:6362–7.
Scrucca L, Santucci A, Aversa F. Competing risk analysis using R: an easy guide for clinicians. Bone Marrow Transplant. 2007;40:381–7.
Acknowledgements
This work was partially supported by grants from the National Natural Science Foundation of China (82070149, 81870109, and 81670135 to X-NG), the Natural Science Foundation of Beijing Municipality (7202191 to X-NG).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Chen, Z., Shao, YL., Wang, LL. et al. YTHDF2 is a potential target of AML1/ETO-HIF1α loop-mediated cell proliferation in t(8;21) AML. Oncogene 40, 3786–3798 (2021). https://doi.org/10.1038/s41388-021-01818-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-01818-1
This article is cited by
-
Small molecule inhibitors targeting m6A regulators
Journal of Hematology & Oncology (2024)
-
A novel AML1-ETO/FTO positive feedback loop promotes leukemogenesis and Ara-C resistance via stabilizing IGFBP2 in t(8;21) acute myeloid leukemia
Experimental Hematology & Oncology (2024)
-
Metabolic dependencies of acute myeloid leukemia stem cells
International Journal of Hematology (2024)
-
m6A methylation: a process reshaping the tumour immune microenvironment and regulating immune evasion
Molecular Cancer (2023)
-
HIF1α-mediated transactivation of WTAP promotes AML cell proliferation via m6A-dependent stabilization of KDM4B mRNA
Leukemia (2023)