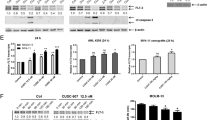
Abstract
FLT3 inhibitors (FLT3i) are widely used for the treatment of acute myeloid leukemia (AML), but adaptive and acquired resistance remains a primary challenge. Inhibitors simultaneously blocking adaptive and acquired resistance are highly demanded. Here, we observed the potential of CHK1 inhibitors to synergistically improve the therapeutic effect of FLT3i in FLT3-mutated AML cells. Notably, the combination overcame adaptive resistance. The simultaneous targeting of FLT3 and CHK1 kinases may overcome acquired and adaptive resistance. A dual FLT3/CHK1 inhibitor 30 with a good oral PK profile was identified. Mechanistic studies indicated that 30 inhibited FLT3 and CHK1, downregulated the c-Myc pathway and further activated the p53 pathway. Functional studies showed that 30 was more selective against cells with various FLT3 mutants, overcame adaptive resistance in vitro, and effectively inhibited resistant FLT3-ITD AML in vivo. Moreover, 30 showed favorable druggability without significant blood toxicity or myelosuppression and exhibited a good oral PK profile with a T1/2 over 12 h in beagles. These findings support the targeting of FLT3 and CHK1 as a novel strategy for overcoming adaptive and acquired resistance to FLT3i therapy in AML and suggest 30 as a potential clinical candidate.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
For original data and reagents, please contact the corresponding author.
References
Sami SA, Darwish NHE, Barile ANM, Mousa SA. Current and Future Molecular Targets for Acute Myeloid Leukemia Therapy. Curr Treat Options Oncol. 2020;21:3.
Burnett A, Wetzler M, Lowenberg B. Therapeutic advances in acute myeloid leukemia. J Clin Oncol. 2011;29:487–94.
Daver N, Schlenk RF, Russell NH, Levis MJ. Targeting FLT3 mutations in AML: review of current knowledge and evidence. Leukemia. 2019;33:299–312.
Wang A, Li X, Chen C, Wu H, Qi Z, Hu C, et al. Discovery of 1-(4-(4-Amino-3-(4-(2-morpholinoethoxy)phenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl)phenyl)-3-(5-(tert-butyl)isoxazol-3-yl)urea (CHMFL-FLT3-213) as a Highly Potent Type II FLT3 Kinase Inhibitor Capable of Overcoming a Variety of FLT3 Kinase Mutants in FLT3-ITD Positive AML. J Medicinal Chem. 2017;60:8407–24.
Nakao M, Yokota S, Iwai T, Kaneko H, Horiike S, Kashima K, et al. Internal tandem duplication of the flt3 gene found in acute myeloid leukemia. Leukemia. 1996;10:1911–8.
Breitenbuecher F, Schnittger S, Grundler R, Markova B, Carius B, Brecht A, et al. Identification of a novel type of ITD mutations located in nonjuxtamembrane domains of the FLT3 tyrosine kinase receptor. Blood. 2009;113:4074–7.
Wang Z, Cai J, Ren J, Chen Y, Wu Y, Cheng J, et al. Discovery of a Potent FLT3 Inhibitor (LT-850-166) with the Capacity of Overcoming a Variety of FLT3 Mutations. J Med Chem. 2021;64:14664–701.
Weisberg E, Barrett R, Liu Q, Stone R, Gray N, Griffin JD. FLT3 inhibition and mechanisms of drug resistance in mutant FLT3-positive AML. Drug Resist Updat. 2009;12:81–89.
Antar AI, Otrock ZK, Jabbour E, Mohty M, Bazarbachi A. FLT3 inhibitors in acute myeloid leukemia: ten frequently asked questions. Leukemia. 2020;34:682–96.
Wang Y, Zhi Y, Jin Q, Lu S, Lin G, Yuan H, et al. Discovery of 4-((7H-Pyrrolo[2,3-d]pyrimidin-4-yl)amino)-N-(4-((4-methylpiperazin-1-yl)methyl)phenyl)-1H-pyrazole-3-carboxamide (FN-1501), an FLT3- and CDK-Kinase Inhibitor with Potentially High Efficiency against Acute Myelocytic Leukemia. J Medicinal Chem. 2018;61:1499–518.
Kennedy VE, Smith CC. FLT3 Mutations in Acute Myeloid Leukemia: Key Concepts and Emerging Controversies. Front Oncol. 2020;10:612880.
Eguchi M, Minami Y, Kuzume A, Chi S. Mechanisms Underlying Resistance to FLT3 Inhibitors in Acute Myeloid Leukemia. Biomedicines. 2020;8:245.
Alvarado Y, Kantarjian HM, Luthra R, Ravandi F, Borthakur G, Garcia-Manero G, et al. Treatment with FLT3 inhibitor in patients with FLT3-mutated acute myeloid leukemia is associated with development of secondary FLT3-tyrosine kinase domain mutations. Cancer. 2014;120:2142–9.
Bagrintseva K, Schwab R, Kohl TM, Schnittger S, Eichenlaub S, Ellwart JW, et al. Mutations in the tyrosine kinase domain of FLT3 define a new molecular mechanism of acquired drug resistance to PTK inhibitors in FLT3-ITD-transformed hematopoietic cells. Blood. 2004;103:2266–75.
Joshi SK, Sharzehi S, Pittsenbarger J, Bottomly D, Tognon CE, McWeeney SK, et al. A noncanonical FLT3 gatekeeper mutation disrupts gilteritinib binding and confers resistance. Am J Hematol. 2021;96:E226–E229.
Pauwels D, Sweron B, Cools J. The N676D and G697R mutations in the kinase domain of FLT3 confer resistance to the inhibitor AC220. Haematologica. 2012;97:1773–4.
Melgar K, Walker MM, Jones LM, Bolanos LC, Hueneman K, Wunderlich M, et al. Overcoming adaptive therapy resistance in AML by targeting immune response pathways. Sci Transl Med. 2019;11:eaaw8828.
Bruner JK, Ma HS, Li L, Qin ACR, Rudek MA, Jones RJ, et al. Adaptation to TKI Treatment Reactivates ERK Signaling in Tyrosine Kinase-Driven Leukemias and Other Malignancies. Cancer Res. 2017;77:5554–63.
Lindblad O, Cordero E, Puissant A, Macaulay L, Ramos A, Kabir NN, et al. Aberrant activation of the PI3K/mTOR pathway promotes resistance to sorafenib in AML. Oncogene. 2016;35:5119–31.
Long J, Jia MY, Fang WY, Chen XJ, Mu LL, Wang ZY, et al. FLT3 inhibition upregulates HDAC8 via FOXO to inactivate p53 and promote maintenance of FLT3-ITD+ acute myeloid leukemia. Blood. 2020;135:1472–83.
Pei HZ, Zhuang X, Yang M, Guo Y, Chang Z, Zhang D, et al. FLT3 Inhibitors Induce p53 Instability Due to Increased Interaction between p53 and MDM2 By Inhibiting STAT5 Phosphorylation in Acute Myeloid Leukemia. Blood. 2021;138:3333.
Pei HZ, Guo Y, Lu B, Chang Z, Zhang D, Yu L, et al. FLT3 Inhibitors Induce Instability of p53 By Mir-181 Mediated Downregulation of Deubiquitinase YOD1 in Acute Myeloid Leukemia. Blood. 2020;136:6–7.
Zhang Y, Hunter T. Roles of Chk1 in cell biology and cancer therapy. Int J Cancer. 2014;134:1013–23.
Cavelier C, Didier C, Prade N, Mansat-De Mas V, Manenti S, Recher C, et al. Constitutive activation of the DNA damage signaling pathway in acute myeloid leukemia with complex karyotype: potential importance for checkpoint targeting therapy. Cancer Res. 2009;69:8652–61.
David L, Fernandez-Vidal A, Bertoli S, Grgurevic S, Lepage B, Deshaies D, et al. CHK1 as a therapeutic target to bypass chemoresistance in AML. Sci Signal. 2016;9:ra90.
Zhang Y, Yuan L. Fms-like tyrosine kinase 3-internal tandem duplications epigenetically activates checkpoint kinase 1 in acute myeloid leukemia cells. Sci Rep. 2021;11:13236.
Yuan LL, Green A, David L, Dozier C, Recher C, Didier C, et al. Targeting CHK1 inhibits cell proliferation in FLT3-ITD positive acute myeloid leukemia. Leuk Res. 2014;38:1342–9.
Tong L, Song P, Jiang K, Xu L, Jin T, Wang P, et al. Discovery of (R)-5-((5-(1-methyl-1H-pyrazol-4-yl)-4-(methylamino)pyrimidin-2-yl)amino)-3-(pipe ridin-3-yloxy)picolinonitrile, a novel CHK1 inhibitor for hematologic malignancies. Eur J Med Chem. 2019;173:44–62.
Jin T, Wang P, Long X, Jiang K, Song P, Wu W, et al. Design, Synthesis, and Biological Evaluation of Orally Bioavailable CHK1 Inhibitors Active against Acute Myeloid Leukemia. ChemMedChem. 2021;16:1477–87.
Green AS, Maciel TT, Hospital MA, Yin C, Mazed F, Townsend EC, et al. Pim kinases modulate resistance to FLT3 tyrosine kinase inhibitors in FLT3-ITD acute myeloid leukemia. Sci Adv. 2015;1:e1500221.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102:15545–50.
Lopez JS, Banerji U. Combine and conquer: challenges for targeted therapy combinations in early phase trials. Nat Rev Clin Oncol. 2017;14:57–66.
Jin T, Xu L, Wang P, Hu X, Zhang R, Wu Z, et al. Discovery and Development of a Potent, Selective, and Orally Bioavailable CHK1 Inhibitor Candidate: 5-((4-((3-Amino-3-methylbutyl)amino)-5-(trifluoromethyl)pyrimidin-2-yl)amino)picolinonitrile. J Medicinal Chem. 2021;64:15069–90.
Tong L, Wang P, Li X, Dong X, Hu X, Wang C, et al. Identification of 2-Aminopyrimidine Derivatives as FLT3 Kinase Inhibitors with High Selectivity over c-KIT. J Med Chem. 2022;65:3229–48.
Galanis A, Levis M. Inhibition of c-Kit by tyrosine kinase inhibitors. Haematologica. 2015;100:e77–79.
Wang GT, Li G, Mantei RA, Chen Z, Kovar P, Gu W, et al. 1-(5-Chloro-2-alkoxyphenyl)-3-(5-cyanopyrazin-2-yl)ureas [correction of cyanopyrazi] as potent and selective inhibitors of Chk1 kinase: synthesis, preliminary SAR, and biological activities. J Med Chem. 2005;48:3118–21.
Jiang KL, Tong LX, Wang T, Wang HL, Hu XB, Xu GY, et al. Downregulation of c-Myc expression confers sensitivity to CHK1 inhibitors in hematologic malignancies. Acta Pharm Sin. 2022;43:220–8.
Takahashi S. Downstream molecular pathways of FLT3 in the pathogenesis of acute myeloid leukemia: biology and therapeutic implications. J Hematol Oncol. 2011;4:13.
Ando K, Nakamura Y, Nagase H, Nakagawara A, Koshinaga T, Wada S, et al. Co-Inhibition of the DNA Damage Response and CHK1 Enhances Apoptosis of Neuroblastoma Cells. Int J Mol Sci. 2019;20:3700.
Larizza L, Magnani I, Beghini A. The Kasumi-1 cell line: a t(8;21)-kit mutant model for acute myeloid leukemia. Leuk lymphoma. 2005;46:247–55.
Piloto O, Wright M, Brown P, Kim KT, Levis M, Small D. Prolonged exposure to FLT3 inhibitors leads to resistance via activation of parallel signaling pathways. Blood. 2007;109:1643–52.
Chandarlapaty S. Negative feedback and adaptive resistance to the targeted therapy of cancer. Cancer Disco. 2012;2:311–9.
Smith CC, Paguirigan A, Jeschke GR, Lin KC, Massi E, Tarver T, et al. Heterogeneous resistance to quizartinib in acute myeloid leukemia revealed by single-cell analysis. Blood. 2017;130:48–58.
Joshi SK, Nechiporuk T, Bottomly D, Piehowski PD, Reisz JA, Pittsenbarger J, et al. The AML microenvironment catalyzes a stepwise evolution to gilteritinib resistance. Cancer Cell. 2021;39:999–1014.e1018.
Kadia TM, Jain P, Ravandi F, Garcia-Manero G, Andreef M, Takahashi K, et al. TP53 mutations in newly diagnosed acute myeloid leukemia: Clinicomolecular characteristics, response to therapy, and outcomes. Cancer. 2016;122:3484–91.
Levine AJ. p53: 800 million years of evolution and 40 years of discovery. Nat Rev Cancer. 2020;20:471–80.
Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53.
Yang M, Pan Z, Huang K, Busche G, Liu H, Gohring G, et al. A unique role of p53 haploinsufficiency or loss in the development of acute myeloid leukemia with FLT3-ITD mutation. Leukemia. 2022;36:675–86.
Sasca D, Hahnel PS, Szybinski J, Khawaja K, Kriege O, Pante SV, et al. SIRT1 prevents genotoxic stress-induced p53 activation in acute myeloid leukemia. Blood. 2014;124:121–33.
Seipel K, Marques MAT, Sidler C, Mueller BU, Pabst T. MDM2- and FLT3-inhibitors in the treatment of FLT3-ITD acute myeloid leukemia, specificity and efficacy of NVP-HDM201 and midostaurin. Haematologica. 2018;103:1862–72.
Andreeff M, Zhang W, Kumar P, Zernovak O, Daver NG, Isoyama T, et al. Synergistic Anti-Leukemic Activity with Combination of FLT3 Inhibitor Quizartinib and MDM2 Inhibitor Milademetan in FLT3-ITD Mutant/p53 Wild-Type Acute Myeloid Leukemia Models. Blood. 2018;132:2720.
Acknowledgements
We thank all the patients whose samples used in the study and their families. We thank Jianyang Pan (Research and Service Center, College of Pharmaceutical Sciences, Zhejiang University) for performing NMR spectrometry for structure elucidation. We also thank Huazhou Ying (College of Pharmaceutical Sciences, Zhejiang University) for performing mass spectrometry for structure elucidation.
Funding
This work was supported by grants from the Guangdong High-level new R&D Institute (2019B090904008) and the Guangdong High-level Innovative Research Institute (2021B0909050003), the key project of Zhejiang Provincial Natural Science Foundation of China (LZ21H300001), the National Natural Science Foundation of China (81821005, 82204179), the Science and Technology Commission of Shanghai Municipality (18431907100, 19430750100).
Author information
Authors and Affiliations
Contributions
KLJ, XML, CW, XBH, TL, YBZ and JL conceived and designed the study. KLJ, CW and YTT performed in vitro cellular experiments, analyzed the data and interpreted the results. PPW performed in vitro enzymatic assays, analyzed the data and interpreted the results. XBH, BJC and RZG performed in vivo experiments, analyzed the data and interpreted the results. XML, LXT, TTJ and TL designed and synthesized the compounds, analyzed the data and interpreted the results. HLW and YBH analyzed RNAseq data. TW and JMY provided patients samples. KLJ, XML, CW, TL and YBZ wrote, reviewed, and/or revised the manuscript. JMY, TL, YBZ, and JL contributed to the study supervision.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jiang, K., Li, X., Wang, C. et al. Dual inhibition of CHK1/FLT3 enhances cytotoxicity and overcomes adaptive and acquired resistance in FLT3-ITD acute myeloid leukemia. Leukemia 37, 539–549 (2023). https://doi.org/10.1038/s41375-022-01795-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-022-01795-8
This article is cited by
-
Therapeutic advances of targeting receptor tyrosine kinases in cancer
Signal Transduction and Targeted Therapy (2024)
-
Recent advancements in biomarkers, therapeutics, and associated challenges in acute myeloid leukemia
Annals of Hematology (2024)