Abstract
Background/objectives
Obesity, defined as excessive fat accumulation that represents a health risk, is increasing in adults and children, reaching global epidemic proportions. Body mass index (BMI) correlates with body fat and future health risk, yet differs in prediction by fat distribution, across populations and by age. Nonetheless, few genetic studies of BMI have been conducted in ancestrally diverse populations. Gene expression association with BMI was assessed in the Multi-Ethnic Study of Atherosclerosis (MESA) in four self-identified race and ethnicity (SIRE) groups to identify genes associated with obesity.
Subjects/methods
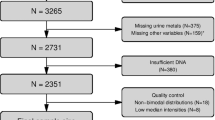
RNA-sequencing was performed on 1096 MESA participants (37.8% white, 24.3% Hispanic, 28.4% African American, and 9.5% Chinese American) and linear models were used to assess the association of expression from each gene for its effect on BMI, adjusting for age, sex, sequencing center, study site, five expression and four genetic principal components in each self-identified race group. Sample-size-weighted meta-analysis was performed to identify genes with BMI-associated expression across ancestry groups.
Results
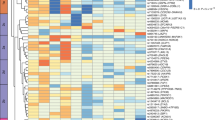
Within individual SIRE groups, there were zero to three genes whose expression is significantly (p < 1.97 × 10–6) associated with BMI. Across all groups, 45 genes were identified by meta-analysis whose expression was significantly associated with BMI, explaining 29.7% of BMI variation. The 45 genes are expressed in a variety of tissues and cell types and are enriched for obesity-related processes including erythrocyte function, oxygen binding and transport, and JAK-STAT signaling.
Conclusions
We have identified genes whose expression is significantly associated with obesity in a multi-ethnic cohort. We have identified novel genes associated with BMI as well as confirmed previously identified genes from earlier genetic analyses. These novel genes and their biological pathways represent new targets for understanding the biology of obesity as well as new therapeutic intervention to reduce obesity and improve global public health.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
MESA data are available through the application to dbGaP. Phenotypes are available in MESA study accession phs000209.v13.p3, and transcriptomic data has been deposited and will become available through the TOPMed MESA study accession phs001416.v2.p1.
References
GBD 2015 Obesity Collaborators. Health effects of overweight and obesity in 195 countries over 25 years. N Engl J Med. 2017;377:13–27.
World Health Organization. Global health estimates 2020: deaths by cause, age, sex, by country and by region, 2000–2019. 2020.
Hoffmann TJ, Choquet H, Yin J, Banda Y, Kvale MN, Glymour M, et al. A large multiethnic genome-wide association study of adult body mass index identifies novel loci. Genetics. 2018;210:499–515.
Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206.
Turcot V, Lu Y, Highland HM, Schurmann C, Justice AE, Fine RS, et al. Protein-altering variants associated with body mass index implicate pathways that control energy intake and expenditure in obesity. Nat Genet. 2018;50:26–41.
Graff M, Scott RA, Justice AE, Young KL, Feitosa MF, Barata L, et al. Genome-wide physical activity interactions in adiposity – a meta-analysis of 200,452 adults. PLoS Genet. 2017;13:e1006528.
Kichaev G, Bhatia G, Loh PR, Gazal S, Burch K, Freund MK, et al. Leveraging polygenic functional enrichment to improve GWAS power. Am J Hum Genet. 2019;104:65–75.
Pulit SL, Stoneman C, Morris AP, Wood AR, Glastonbury CA, Tyrrell J, et al. Meta-analysis of genome-wide association studies for body fat distribution in 694 649 individuals of European ancestry. Hum Mol Genet. 2019;28:166–74.
Zhu Z, Guo Y, Shi H, Liu CL, Panganiban RA, Chung W, et al. Shared genetic and experimental links between obesity-related traits and asthma subtypes in UK Biobank. J Allergy Clin Immunol. 2020;145:537–49.
Winkler TW, Justice AE, Graff M, Barata L, Feitosa MF, Chu S, et al. The influence of age and sex on genetic associations with adult body size and shape: a large-scale genome-wide interaction study. PLoS Genet. 2015;11:e1005378.
Yengo L, Sidorenko J, Kemper KE, Zheng Z, Wood AR, Weedon MN, et al. Meta-analysis of genome-wide association studies for height and body mass index in approximately 700000 individuals of European ancestry. Hum Mol Genet. 2018;27:3641–9.
Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nature Genetics. 2010;42:937–48.
Young KL, Graff M, Fernandez-Rhodes L, North KE. Genetics of obesity in diverse populations. Curr Diab Rep. 2018;18:145.
Bild DE. Multi-Ethnic Study of Atherosclerosis: objectives and design. Am J Epidemiol. 2002;156:871–81.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21.
DeLuca DS, Levin JZ, Sivachenko A, Fennell T, Nazaire MD, Williams C, et al. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics. 2012;28:1530–2.
Frankish A, Diekhans M, Jungreis I, Lagarde J, Loveland Jane E, Mudge JM, et al. Gencode 2021. Nucleic Acids Res. 2021;49:D916–23.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Sofer T, Kurniansyah N, Aguet F, Ardlie K, Durda P, Nickerson DA, et al. Benchmarking association analyses of continuous exposures with RNA-seq in observational studies. Brief Bioinform. 2021;22:bbab194.
van Iterson M, van Zwet EW, Heijmans BT. Controlling bias and inflation in epigenome- and transcriptome-wide association studies using the empirical null distribution. Genome Biol. 2017;18:19.
Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1.
Cochran WG. The combination of estimates from different experiments. Biometrics. 1954;10:101–29.
Higgins JPT, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–58.
Zhang D. rsq: R-squared and related measures. R Package. 2022.
Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47:D1005–12.
Loh P-R, Genovese G, Handsaker RE, Finucane HK, Reshef YA, Palamara PF, et al. Insights into clonal haematopoiesis from 8,342 mosaic chromosomal alterations. Nature. 2018;559:350–5.
Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (Camb). 2021;2:100141.
Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–7.
Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30.
Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49:D545–51.
Ye Y, Doak TG. A parsimony approach to biological pathway reconstruction/inference for genomes and metagenomes. PLoS Comput Biol. 2009;5:e1000465.
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9.
Gene Ontology Consortium The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res. 2021;49:D325–34.
Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, et al. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–5.
Paccosi S, Pala L, Cresci B, Silvano A, Cecchi M, Caporale R, et al. Insulin resistance and obesity affect monocyte-derived dendritic cell phenotype and function. Diabetes Res Clin Pract. 2020;170:108528.
Mishra J, Verma RK, Alpini G, Meng F, Kumar N. Role of Janus Kinase 3 in predisposition to obesity-associated metabolic syndrome. J Biol Chem. 2015;290:29301–12.
Ghadami M, Tomita HA, Najafi MT, Damavandi E, Farahvash MS, Yamada K, et al. Bardet-Biedl syndrome type 3 in an Iranian family: clinical study and confirmation of disease localization. Am J Med Genet. 2000;94:433–7.
Young TL, Woods MO, Parfrey PS, Green JS, O’Leary E, Hefferton D, et al. Canadian Bardet-Biedl syndrome family reduces the critical region of BBS3 (3p) and presents with a variable phenotype. Am J Med Genet. 1998;78:461–7.
Mazzarella L, Botteri E, Matthews A, Gatti E, Di Salvatore D, Bagnardi V, et al. Obesity is a risk factor for acute promyelocytic leukemia: evidence from population and cross-sectional studies and correlation with FLT3 mutations and polyunsaturated fatty acid metabolism. Haematologica. 2020;105:1559–66.
Díaz-Rúa R, Palou A, Oliver P. Cpt1agene expression in peripheral blood mononuclear cells as an early biomarker of diet-related metabolic alterations. Food Nutr Res. 2016;60:33554.
Orellana-Gavaldà JM, Herrero L, Malandrino MI, Pañeda A, Sol Rodríguez-Peña M, Petry H, et al. Molecular therapy for obesity and diabetes based on a long-term increase in hepatic fatty-acid oxidation. Hepatology. 2011;53:821–32.
Hauffe R, Stein V, Chudoba C, Flore T, Rath M, Ritter K, et al. GPx3 dysregulation impacts adipose tissue insulin receptor expression and sensitivity. JCI Insight. 2020;5:e136283.
Langhardt J, Flehmig G, Klöting N, Lehmann S, Ebert T, Kern M, et al. Effects of weight loss on glutathione Peroxidase 3 serum concentrations and adipose tissue expression in human obesity. Obesity Facts. 2018;11:475–90.
Paulo E, Wu D, Hecker P, Zhang Y, Wang B. Adipocyte HDAC4 activation leads to beige adipocyte expansion and reduced adiposity. J Endocrinol. 2018;239:153–65.
Bereswill S, Abu-Farha M, Tiss A, Abubaker J, Khadir A, Al-Ghimlas F, et al. Proteomics analysis of human obesity reveals the epigenetic factor HDAC4 as a potential target for obesity. PLoS One. 2013;8:e75342.
Netea MG, Joosten LAB, Lewis E, Jensen DR, Voshol PJ, Kullberg BJ, et al. Deficiency of interleukin-18 in mice leads to hyperphagia, obesity and insulin resistance. Nat Med. 2006;12:650–6.
Ennequin G, Boisseau N, Caillaud K, Chavanelle V, Etienne M, Li X, et al. Neuregulin 1 affects leptin levels, food intake and weight gain in normal-weight, but not obese, db/db mice. Diabetes Metab. 2015;41:168–72.
Val CH, de Oliveira MC, Lacerda DR, Barroso A, Batista NV, Menezes-Garcia Z, et al. SOCS2 modulates adipose tissue inflammation and expansion in mice. J Nutr Biochem. 2020;76:108304.
Li J, Diao B, Guo S, Huang X, Yang C, Feng Z, et al. VSIG4 inhibits proinflammatory macrophage activation by reprogramming mitochondrial pyruvate metabolism. Nat Commun. 2017;8:1322.
Gurzov EN, Stanley WJ, Pappas EG, Thomas HE, Gough DJ. The JAK/STAT pathway in obesity and diabetes. FEBS J. 2016;283:3002–15.
Ge D, Gooljar SB, Kyriakou T, Collins LJ, Swaminathan R, Snieder H, et al. Association of common JAK2 variants with body fat, insulin sensitivity and lipid profile. Obesity (Silver Spring). 2008;16:492–6.
Penas-Steinhardt A, Tellechea ML, Gomez-Rosso L, Brites F, Frechtel GD, Poskus E. Association of common variants in JAK2 gene with reduced risk of metabolic syndrome and related disorders. BMC Med Genet. 2011;12:166.
Dodington DW, Desai HR, Woo M. JAK/STAT – emerging players in metabolism. Trends Endocrinol Metab. 2018;29:55–65.
Messaoudi I, Handu M, Rais M, Sureshchandra S, Park BS, Fei SS, et al. Long-lasting effect of obesity on skeletal muscle transcriptome. BMC Genomics. 2017;18:411.
Chen J-J, London IM. Hemin enhances the differentiation of mouse 3T3 cells to adipocytes. Cell. 1981;26:117–22.
Hazegh K, Fang F, Bravo MD, Tran JQ, Muench MO, Jackman RP, et al. Blood donor obesity is associated with changes in red blood cell metabolism and susceptibility to hemolysis in cold storage and in response to osmotic and oxidative stress. Transfusion. 2020;61:435–48.
Samocha-Bonet D, Lichtenberg D, Tomer A, Deutsch V, Mardi T, Goldin Y, et al. Enhanced erythrocyte adhesiveness/aggregation in obesity corresponds to low-grade inflammation. Obes Res. 2003;11:403–7.
Moll M, Boueiz A, Ghosh AJ, Saferali A, Lee S, Xu Z, et al. Development of a blood-based transcriptional risk score for chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2022;205:161–70.
Loos RJF, Yeo GSH. The genetics of obesity: from discovery to biology. Nat Rev Genet. 2022;23:120–33.
Banks AS, Davis SM, Bates SH, Myers MG. Activation of downstream signals by the long form of the leptin receptor. J Biol Chem. 2000;275:14563–72.
Acknowledgements
Molecular data for the Trans-Omics in Precision Medicine (TOPMed) program was supported by the National Heart, Lung and Blood Institute (NHLBI). Genome sequencing for “NHLBI TOPMed: Multi-Ethnic Study of Atherosclerosis (MESA)” (phs001416.v1.p1) was performed at the Broad Institute of MIT and Harvard (3U54HG003067-13S1). Centralized read mapping and genotype calling, along with variant quality metrics and filtering were provided by the TOPMed Informatics Research Center (3R01HL-117626-02S1 and HHSN268201800002I). Phenotype harmonization, data management, sample-identity QC, and general study coordination, were provided by the TOPMed Data Coordinating Center (3R01HL-120393-02S1), and TOPMed MESA Multi-Omics (HHSN2682015000031/HHSN26800004). The MESA projects are conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with MESA investigators. Support for the Multi-Ethnic Study of Atherosclerosis (MESA) projects are conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with MESA investigators. Support for MESA is provided by contracts 75N92020D00001, HHSN268201500003I, N01-HC-95159, 75N92020D00005, N01-HC-95160, 75N92020D00002, N01-HC-95161, 75N92020D00003, N01-HC-95162, 75N92020D00006, N01-HC-95163, 75N92020D00004, N01-HC-95164, 75N92020D00007, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-TR-000040, UL1-TR-001079, UL1-TR-001420, UL1TR001881, DK063491, and R01HL105756. The authors thank the other investigators, the staff, and the participants of the MESA study for their valuable contributions. A full list of participating MESA investigators and institutes can be found at http://www.mesa-nhlbi.org.
Author information
Authors and Affiliations
Contributions
LBV and IRK performed all statistical analysis, data visualization, and drafted the manuscript. LAL, KF, and EML contributed to conceptualizing analyses and critical editing of the manuscript. JDS, SG, and NG performed RNA-Sequencing. FA and KA processed RNA-Seq data to create the final MESA transcriptomics dataset. TWB provided advice and support as a member of the TOPMed Informatics Research Center. JD provided advice and support as a member of the MESA Multi-omics Adiposity Working Group. PD, RPT, YL, WCJ, SSR, JIR, and KDT designed the RNA-Seq study in MESA. All authors critically reviewed and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
FA is an employee and shareholder of Illumina, Inc. The remaining authors have no competing interests to disclose.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Vargas, L.B., Lange, L.A., Ferrier, K. et al. Gene expression associations with body mass index in the Multi-Ethnic Study of Atherosclerosis. Int J Obes 47, 109–116 (2023). https://doi.org/10.1038/s41366-022-01240-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41366-022-01240-x