Abstract
Until recently, Nurr1 (NR4A2) was known as an orphan nuclear receptor without a canonical ligand-binding domain, featuring instead a narrow and tight cavity for small molecular ligands to bind. In-depth characterization of its ligand-binding pocket revealed that it is highly dynamic, with its structural conformation changing more than twice on the microsecond-to-millisecond timescale. This observation suggests the possibility that certain ligands are able to squeeze into this narrow space, inducing a conformational change to create an accessible cavity. The cocrystallographic structure of Nurr1 bound to endogenous ligands such as prostaglandin E1/A1 and 5,6-dihydroxyindole contributed to clarifying the crucial roles of Nurr1 and opening new avenues for therapeutic interventions for neurodegenerative and/or inflammatory diseases related to Nurr1. This review introduces novel endogenous and synthetic Nurr1 agonists and discusses their potential effects in Nurr1-related diseases.
Similar content being viewed by others
Introduction
In general, a nuclear receptor is a ligand-regulated transcription factor that regulates downstream gene transcription in the nucleus in response to binding to its specific ligand(s)1. Nuclear receptors with no known specific ligand(s) are referred to as orphan nuclear receptors2. Until recently, the transcription factor Nurr1, which belongs to the NR4A subfamily, was considered an orphan nuclear receptor. The NR4A subfamily of nuclear receptors includes Nur77 (NR4A1), Nurr1 (NR4A2), and Nor1 (NR4A3)3,4. All family members have a ligand-binding domain (LBD) that shares sequence homology with the LBDs of canonical nuclear receptors5,6. Nevertheless, it was widely believed that Nurr1 is constitutively active in a ligand-independent manner5,6. In fact, crystal structural analysis revealed that the Nurr1-LBD lacks a classical binding pocket for coactivators and/or ligands due to the tight packing of bulky hydrophobic side chain residues7. However, recent NMR solution and HDX-MS characterizations revealed that the Nurr1-LBD is highly dynamic with high solvent accessibility, showing a change in structural conformation more than twice on the microsecond-to-millisecond timescale8. Numerous researchers are gradually identifying and validating the endogenous and synthetic ligands that activate Nurr1. Furthermore, recent characterization of Nurr1’s endogenous ligands has aroused further interest in the development of Nurr1 agonists and therapeutic drugs for diseases related to the receptor9,10. Therefore, this review focuses on putative ligands known to induce/enhance Nurr1 transcriptional activity.
In the midbrain, Nurr1 is indispensable for the differentiation, maturation, and maintenance of midbrain dopaminergic neuron clusters (referred to as A9 and A10), which reside in the ventral tegmental area (VTA) and the substantia nigra pars compacta (SNpc), respectively11,12. Nurr1 is known to regulate the transcription of dopamine-related crucial genes, including tyrosine hydroxylase (TH), dopamine transporter (DAT), vesicular monoamine transporter 2 (VMAT2), and aromatic amino acid decarboxylase (AADC), which significantly influence striatal dopamine levels5,13. To modulate the expression of these genes, Nurr1 binds to the nerve growth factor-induced clone B (NGFI-B) response element specific sequence (NBRE; 5′-AAAGGTCA-3′) as a monomer or to the inverted repeat octanucleotide Nur response element (NurRE; 5′-TGACCTTT-n6-AAAGGTCA-3′) as a homodimer14,15. In addition, Nurr1 can interact with retinoid-X receptor alpha (RXRα), forming a heterodimer that binds to the DR5-specific sequence element (5’-GGTTCACCGAAAGGTCA-3’)14,15. Indeed, Nurr1-deficient mice fail to generate midbrain dopaminergic neurons16, and tamoxifen-induced disruption of Nurr1 in the midbrain of adult mice also results in a progressive reduction in the number of striatal dopaminergic neurons17. Thus, these findings support Nurr1’s critical roles in both the development and maintenance of midbrain dopaminergic neurons.
In addition, Nurr1 is also known to exert neuroprotective effects against neuroinflammation of dopaminergic neurons. The promoter region of Nurr1 contains the transcriptional binding site of cAMP-responsive element-binding protein (CREB), and its interaction induces the upregulation of Nurr1 transcripts. This CREB-mediated Nurr1 upregulation seems to be involved in neuroprotection as a regulatory mechanism downstream of CREB18. Thus, it is possible that G-protein coupled receptors that increase intracellular cAMP levels, such as brain-derived neurotrophic factor (BDNF) and prostanoid receptors, which are involved in the neuroprotective pathways for dopaminergic neurons, are highly associated with Nurr1. In neuroprotective pathways, Nurr1 is known to regulate the expression of various nuclear-encoded mitochondrial genes in dopaminergic neurons17. It was found that a significant reduction in the levels of superoxide dismutase 1 (SOD1), which is one of these mitochondrial genes, is strongly implicated in oxidative stress detoxification in Nurr1-ablated cells17. Furthermore, Nurr1 is known to transcriptionally repress proinflammatory genes in microglia and astrocytes upon exposure to bacterial lipopolysaccharide (LPS), suggesting that it exerts neuroprotection by suppressing neurotoxic proinflammatory genes in the brain during inflammation19,20,21. Taken together, this evidence indicates that Nurr1 transcriptionally promotes the expression of anti-inflammatory genes, represses the expression of proinflammatory genes, and protects dopaminergic neurons.
While Nurr1 is important for the maintenance and protection of dopaminergic neurons, it has been reported that Nurr1 expression in the brains of Parkinson’s disease (PD) patients and in the brains of rodent models of PD induced by forced expression of α-synuclein or exposure to neurotoxins (6-OHDA or MPTP) is downregulated22,23,24. To date, no mechanistic hypothesis has been put forth to explain this reduced expression. Therefore, the development of selective Nurr1 agonists offers the possibility to provide target-based therapeutic interventions for PD and other related diseases.
Bicyclic compounds
Contrary to the existing concept that Nurr1 is constitutively active, several studies have attempted to identify potent Nurr1 agonists from established chemical libraries. Because Nurr1 binds to the promoter region of target genes, cell-based luciferase reporter assays using specific Nurr1-binding sequence(s) have been used for high-throughput screenings.
6-Mercaptopurine was the first Nurr1 agonist identified from a 340,800 compound prototypic library (Fig. 1a)25. 6-Mercaptopurine is commonly used for treating cancer and autoimmune diseases such as acute/chronic leukemia, ulcerative colitis, and Crohn’s disease25,26,27. It was shown to robustly enhance Nurr1 transcriptional activity using the NurRE target sequence in a concentration-dependent manner in CV-1 cells25. In the following year, reports indicated that bicyclic isoxazolopyridinone-based compounds increase Nurr1 reporter gene activity as a homodimer and a heterodimer using the NurRE and DR5 sequence elements, respectively (Fig. 1b)28. The molecular interaction between Nurr1-LBD and these compounds was demonstrated by a radiolabeled binding assay28. Additionally, these isoxazolopyridinones were shown to increase dopamine levels in the substantia nigra (SN) and striatumof OF1 mice at doses of 5 to 30 mg/kg28.
Based on the benzimidazole scaffold, a bicyclic compound of fused benzene and imidazole (Fig. 1c), Dubois and colleagues used combinatorial chemistry and structure-based drug design to develop a potential Nurr1 agonist29. They synthesized 3840 compounds harboring the benzimidazole scaffold and tested for potential agonists using a Nurr1 DNA binding domain (DBD)-containing luciferase construct in MN9D cells. To rule out the possibility of heterodimer activation of Nurr1 and RXRα, they also assessed luciferase activity in the RXRα-overexpressing cell line. Using this reporter assay, they identified three potent Nurr1 agonists active with an EC50 in the nanomolar range (8–70 nM)29. In 2008, the pharmaceutical company Sanofi Aventis introduced the bicyclic-based Nurr1 agonist SA00025, a 2-aryl-6-phenylimidazo[1,2-α]pyridine derivative (Fig. 1d)30. In vivo, oral administration of SA00025 (30 mg/kg) for 32 days protects dopaminergic neurons in a 6-OHDA-induced lesion rat model of PD exacerbated by inflammation31.
Recently, Lesuisse et al. found two additional bicyclic imidazopyridines that act as Nurr1 agonists through compound screening based on the NBRE sequence coupled to a luciferase reporter gene in the neuronal N2A cell line32. The identified imidazopyridine-derived compounds are N-phenyl-6-(pyrid-2-yl)imidazo[1,2-a]pyridine-2-carboxamide (Fig. 1e) and (3-[2-(4-chlorophenyl)imidazo [1,2-a]pyridin-6-yl]phenyl)methanol (Fig. 1f)32. They enhance Nurr1 transcriptional activity using the NBRE target sequence at an EC50 of 1 nM32.
Amodiaquine (AQ) and chloroquine (CQ)
Molecular interactions of AQ/CQ with Nurr1
Using a cell-based luciferase assay system, Kim and colleagues screened a chemical library composed of 960 FDA-approved drugs and identified 3 potential Nurr1 activators, namely, amodiaquine (AQ) (Fig. 1g), chloroquine (CQ) (Fig. 1h), and glafenine (Gla)33. Interestingly, all three compounds share an identical chemical scaffold, 4-amino-7-chloroquinoline, suggesting a specific structure-activity relationship for these Nurr1 activators. Using a luciferase-based reporter assay consisting of the DBD of the yeast transcription factor GAL4 fused to the Nurr1-LBD, AQ and CQ were shown to increase the transcriptional activities of Nurr1-LBD and full-length Nurr1 in the human neuroblastoma cell line SK-N-BE(2)C33. Moreover, using radiolabeled [3H]-CQ, the researchers showed saturable binding to Nurr1-LBD with a dissociation constant (Kd) of 0.27 μM and a maximal binding capacity (Bmax) of 13.9 μM33. Unlabeled AQ competed for the binding of [3H]-CQ to Nurr1-LBD, indicating that AQ/CQ binds to similar Nurr1-LBD residues. Furthermore, nuclear magnetic resonance (NMR) spectroscopy also suggested the existence of molecular interactions between AQ/CQ and Nurr1-LBD. The comparison of Nurr1-LBD spectra for free and AQ-bound showed that multiple residues were shifted in the helix α2 region (His402, Ile403, Gln404, Gln405, Asp408, and Leu409) and the helix α11 region (Val468, Tyr575, and Asp580)33. CQ-bound spectra showed perturbed residues mostly in the helix α4 (Ser441) and the helix α12 region (Ile573, Ala586, Ile588, Lys590, Leu593, Asp594, Thr595, Leu596, and Phe598) of Nurr1-LBD34.
Nurr1-dependent biological effect of AQ/CQ
Malaria is a serious tropical disease associated with high fever, chills, and flu-like symptoms and is typically transmitted by Plasmodium parasites through mosquito bites35. Originally, AQ/CQ was developed as anti-malaria drugs, and they are still commonly used to treat malaria36. Since AQ/CQ is known as active Nurr1 ligands, several studies have demonstrated the therapeutic effects of AQ/CQ based on Nurr1 modulation in several neurodegenerative and inflammatory diseases33,34.
The biological effects of AQ/CQ were demonstrated in the dopaminergic system. In cultured rat dopaminergic neurons, exposure to AQ was shown to significantly enhance the mRNA levels of Nurr1-target genes such as TH, DAT, VMAT, and AADC in a concentration-dependent manner33. Furthermore, this study revealed that CQ had neuroprotective effects against the neurotoxin 6-OHDA both in vitro and in vivo, resulting in significant improvements in PD-like motor behaviors in 6-OHDA-lesioned rats33. Hedya et al., using the rotenone rat model of PD, showed that the less toxic derivative hydroxychloroquine (HCQ) exerts an anti-inflammatory effect associated with increased Nurr1 expression, reduced activity of GSK-3β, and diminished expression levels of inflammatory mediators (NF-κB, TNF-α, and IL-1β)37. Consequently, treatment with HCQ resulted in amelioration of rotenone-induced impaired motor behaviors37.
Since Nurr1 is highly expressed in hippocampal neurons as well, Moon and colleagues attempted to clarify the function of Nurr1 and the effect of AQ in Alzheimer’s disease (AD), which is characterized by memory loss (dementia) caused by neuronal loss in the hippocampus starting in the early stage of disease38,39. By overexpressing and silencing Nurr1 in the hippocampal area, the researchers demonstrated that Nurr1 ameliorates AD‐related pathological symptoms, including Aβ‐plaque deposition, in 5XFAD mice38. In line with these findings, 5XFAD mice treated with AQ exhibited marked improvements in typical AD pathogenesis, including deposition of Aβ plaques, neuronal loss, impaired adult hippocampal neurogenesis, and cognitive defects38. Further investigation of adult hippocampal neural stem cells determined that AQ increases hippocampal adult neurogenesis through cell cycle progression40.
Recently, Kinoshita et al. demonstrated AQ’s anti-inflammatory effect from a different perspective. Nurr1 is highly expressed in microglia/macrophages and astrocytes in the perihematomal area of the striata of mice with intracerebral hemorrhage (ICH)41. Intraperitoneal injection of AQ (40 mg/kg) after IHC induction prominently attenuated the activation of microglia/macrophages and astrocytes in the perihematomal area, consequently improving impaired motor deficits41. Given that CQ is commonly used to treat autoimmune diseases such as rheumatoid arthritis42,43,44, it is possible that Nurr1 is involved in the differentiation of naïve T cells into regulatory T cells, which are critical to immune homeostasis45,46,47. Indeed, Park et al. found that CQ facilitates differentiation into regulatory T cells via induction of the Foxp3 gene by Nurr134. Intraperitoneal injection of CQ (50 mg/kg) was shown to mitigate colitis inflammation in a dextran sulfate sodium (DSS)-induced colitis mouse model, a valid animal model of inflammatory bowel disease34.
5,6-Dihydroxyindole (DHI)
Molecular interactions of DHI with Nurr1
The biosynthesis and storage of the neurotransmitter dopamine are regulated by several enzymes and cofactors, all of which can control dopamine levels. In dopamine synthesis, TH is a rate-limiting enzyme that converts amino acid tyrosine to L-dopa48,49. Subsequently, L-dopa is converted to dopamine by the enzyme AADC48,49. The resulting dopamine is transported into the synaptic vesicle by VMAT250. Released dopamine is recycled by DAT, which takes it back into the cytosol from the synaptic cleft51.
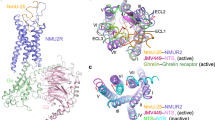
Nurr1’s involvement in dopamine homeostasis includes transcriptional regulation of key factors involved in dopamine synthesis (TH and AADC), vesicle packaging (VMAT2), and dopamine reuptake DAT, which could significantly influence striatal dopamine levels5,13. Therefore, it is possible that Nurr1’s activity in dopamine processing could be tightly regulated by dopamine, its precursors, and/or its metabolites to maintain dopamine homeostasis. In support of this possibility, using biophysical assays and X-ray crystallography, Bruning and colleagues found that the dopamine metabolite 5,6-dihydroxyindole (DHI) directly binds to Nurr1-LBD (Figs. 2a and 3)9. DHI forms a covalent adduct with Cys566 on helix H11 adjacent to the canonical ligand-binding pocket in well-characterized nuclear receptors9. The ligand electron density map and quantum mechanical calculations support the view that a chemical reaction between the Cys566 sulfur atom and the C2 atom of 5,6-indolequinone, the autooxidation product of DHI, leads to adduct formation9. These findings indicate that DHI binds as a quinone to the Cys566 residue of Nurr1. To investigate the effect of DHI on the transcriptional activity of Nurr1, a cell-based luciferase assay using a GAL4 DBD-fused Nurr1 construct was performed, and it was found that DHI induces the transcriptional activity of Nurr1-LBD in JEG3 cells, with 10 μM DHI having a significant effect and 100 μM DHI inducing a 1.6-fold increase in activation over basal levels9.
Biological effects of the interaction of DHI with Nurr1
Little is known about the effect of the recently identified DHI on the dopaminergic system. Under oxidative stress, dopamine can be oxidized into two DHI and 5,6-indolequinone molecules, which are cytotoxic intermediates in the biosynthetic pathway of neuromelanin52. Thus, oxidative stress induces pathological accumulation of cytosolic dopamine in dopaminergic neurons. In particular, the dopamine oxidation product DHI could influence the deposition of α-synuclein aggregates in dopaminergic neurons, a pathogenic feature of PD53. Taken together, this evidence suggests that because excessive dopamine proportionally increases the levels of these reactive metabolites, it is likely that DHI-induced Nurr1 activation regulates the transcription levels of Nurr1 target genes, including those involved in the synthesis, packaging, and reuptake of dopamine.
To verify this possibility, the changes in the transcription levels of several Nurr1 target genes induced by DHI were evaluated in zebrafish. Exposure of zebrafish embryos (3 days post-fertilization) to 100 µM DHI resulted in a significant increase in VMAT2 and DAT transcript levels after 6 h as well as increases in VMAT2 and TH levels after 24 h9. However, the dependence of these changes on Nurr1 expression remains to be elucidated. Nurr1 knockdown and overexpression systems would offer conclusive evidence. In addition, the roles of DHI in the dopaminergic system should be further investigated in rodent models.
Prostaglandin E1 (PGE1) and prostaglandin A1 (PGA1)
Molecular interactions of PGE1 and PGA1 with Nurr1
Prostaglandins (PGs) are bioactive lipid metabolites derived from the metabolism of membrane polyunsaturated fatty acids54. Fatty acids are converted into well-known functional metabolites such as arachidonic acid, eicosapentaenoic acid (EPA), and docosahexaenoic acid (DHA)55. These unsaturated fatty acids have two long hydrocarbon tails, which may allow them to squeeze into the hydrophobic and narrow cavity of the canonical pocket of NR4A-LBDs. In fact, some remarkable studies have highlighted the interaction between unsaturated fatty acids and NR4A-LBDs. For example, a metabolomics-based study of immobilized Nur77-LBD revealed its interaction with unsaturated fatty acids, including arachidonic acid and DHA56. Using a pull-down assay, it was later shown that DHA also interacts with Nurr1-LBD (Fig. 2b)57. Titration of DHA with 15N-labeled Nurr1-LBD showed that DHA binding affects the methyl groups of Leu410, Ile483, and Ile486, which are imbedded in the Nurr1-LBD, resulting in a conformational change in the C-terminal helix 1257. In addition, DHA (12.5 and 50 μM) induced Nurr1 transcriptional activity using the NBRE target sequence in HEK293T and MN9D cells57. Further in-depth analysis revealed that unsaturated fatty acids, including arachidonic acid, linoleic acid, and oleic acid, bind to Nurr1-LBD and induce conformational expansion, which allows more ligands to access the ligand-binding pocket8.
The cyclopentenone PGA2 was the first PG identified as an active ligand of Nor1 and Nur7758,59. Kagaya and colleagues demonstrated a direct interaction between PGA2 and Nor1-LBD using a cell-free Biacore system. Moreover, they showed that PGA2 (10 μM) is a transcriptional activator of Nor1 in NIH3T3 cells58. In addition, incubation of recombinant Nur77 with PGA2-biotin allowed the identification of the PGA2-Nur77 complex through the covalent interaction between PGA2 and the Cys566 residue of Nur7759. In a Nur77-specific reporter assay, treatment with PGA2 caused dose-dependent transcriptional activation of Nur77 but not PGE2 in human bronchial epithelial (NHBE) cells59.
Most recently, an extensive study performed by Rajan et al. in which active compounds were isolated from homogenized mouse tissue extracts identified PGE1 (Fig. 2c) and PGA1 (Fig. 2d) as potent endogenous Nurr1 ligands (Fig. 3)10. Initially, using the DAD of the yeast transcription factor GAL4 fused to the Nurr1-LBD in a human neuroblastoma cell line (SK-N-BE(2)C), the authors found that treatment with several tissue extracts increased Nurr1 transcriptional activity. To isolate the active components from the brain extracts, they performed sequential purification using boiling, acetone precipitation, and ultrafiltration to identify the Nurr1 activity-enriched fractions. Finally, PGE1 and 8-iso PGE1 were identified from several candidate compounds in the final active fraction. In the process of crystallizing the Nurr1-LBD-PGE1 complex, the dehydrated PGE1 metabolite PGA1 was found to form a covalent adduct between the C11 atom of the cyclopentenone ring and the thiol group of the Cys566 residue of Nurr1-LBD. It is likely that crystallization conditions (MES buffer, pH 5.5) lead to the spontaneous dehydration of PGE1 to PGA1. This is supported by several reports showing PGE1 dehydration in acidic/basic environments60,61. An added intriguing feature of Nurr1-LBD binding to PGA1 is that the two long fatty acid tails of PGA1 are squeezed into a hydrophobic space surrounded by helices H4, H11, and H12, inducing the outward movement of helix H12 from the core of Nurr1 at an angle of 21°.
Neuroprotective effects of PGE1 and PGA1 via interaction with Nurr1
As shown in Fig. 4, linoleic acid is a polyunsaturated omega-6 fatty acid with a final carbon-carbon double bond in the n-6 position, whereas α-linolenic acid is an omega-3 fatty acid with three double bonds. Both fatty acids are found in vegetable oils, oily fish, nuts, and seeds at different ratios55. PGE1 is synthesized from dihomo-γ-linolenic acid, which originates from linoleic acid (omega-6)55. PGE2 and PGE3 are enzymatically produced from arachidonic acids originating from linoleic acids and α-linolenic acid-mediated EPA/DHA. PGE1-3 is further converted to PGA1-3 by a nonenzymatic dehydration process62,63. In general, PGEs activate prostaglandin E receptors (EP1-EP4), which subsequently regulate the levels of intracellular second messengers, cAMP, and Ca2+, via G proteins64,65,66,67. Despite extensive investigations across various fields on the roles of PGs, little is known about the actions of PGE1 and PGA1 in dopaminergic neurons where Nurr1 is functionally expressed16.
Double immunohistochemistry for specific EP receptors and TH revealed that EP1/EP2 receptors and EP2/EP3 receptors are selectively expressed in dopaminergic and nondopaminergic neurons in the rat SN68. Unlike EP2- and EP3-selective agonists, a EP1 receptor-specific agonist (17-phenyl trinor PGE2) was shown to exert significant toxic effects on dopaminergic neurons at nanomolar concentrations68. Conversely, antagonism by EP1 receptor blockers (SC-19220 and SC-51089) was shown to protect dopaminergic neurons against 6-OHDA-induced toxicity in rat primary mesencephalic culture68. Moreover, 6-OHDA-induced dopaminergic neuronal death was alleviated by treatment with an EP2 receptor-selective agonist (butaprost)69. Taken together, these findings suggest that the EP2 receptor-mediated cAMP pathway seems to be crucially involved in the survival of dopaminergic neurons.
Based on reporter assays, both PGE1 and PGA1 induce transcriptional activation of Nurr1 in a dose-dependent manner. Interestingly, PGE1-induced Nurr1 activation is inhibited by an EP2-selective antagonist (PF-04418948), whereas PGA1-mediated activation is not. In fact, radiolabeled ligand binding assays with EP2 and Nurr1-LBD proteins using [3H]-PGE1 and [3H]-PGA1 demonstrated that PGE1 interacts with both EP2 and Nurr1 and that PGA1 binds only to Nurr110. Taken together, this evidence indicates that PGE1 regulates the transcriptional activity of Nurr1 via two distinct mechanisms: an EP2-mediated pathway and direct binding to Nurr1-LBD. In contrast, PGA1 covalently binds to the Cys566 residue of Nurr1-LBD and activates its transactivation.
In the midbrain, the PGE1- and PGA1-mediated Nurr1 axis seems to be crucial for protection against dopaminergic neuronal degeneration. PGE1 (3 μM) or PGA1 (5 μM) treatment was shown to protect MN9D and N27-A cells from MPP+-induced toxicity in a Nurr1-dependent manner. PGE1 and PGA1 ameliorated both MPP+- and LPS-induced losses of TH-positive dopaminergic neurons in the nanomolar range in primary dopaminergic neuron-glia cocultures isolated from the rat embryonic ventral mesencephalic area. Moreover, intraperitoneal injection of PGE1 (2 mg/kg) or PGA1 (2 mg/kg) significantly rescued dopaminergic neurons in the SN and the striatum and alleviated impaired motor behaviors in a subchronic MPTP-induced animal model of PD.
Conclusion and perspective
In this review, we introduced recent findings on potent synthetic and endogenous ligands of Nurr1 ligands, a nuclear receptor that has long been referred to as a ligand-independent nuclear receptor (Tables 1, 2). The actions of these activators and ligands strongly support the notion that Nurr1 is not an orphan nuclear receptor that is constitutively active in a ligand-independent manner. Recent in-depth characterization of the structural conformations of Nurr1-LBD and ligand-bound crystal structures further provides molecular and structural insights into the ligand-dependent roles of Nurr1 under physiological and pathological conditions. The dopamine oxidation metabolite DHI has been proposed as a regulatory factor in intracellular dopamine homeostasis through Nurr1 activation (Fig. 3). In addition, PGE1 and PGA1 have been proposed to mediate neuroprotection via activation of Nurr1 in neurodegenerative diseases such as PD and AD. It is likely that extracellular PGE1 regulates the transcriptional activity of Nurr1 via the EP2-mediated pathway, whereas intracellular PGE1/PGA1 bind directly to Nurr1-LBD, activating its transcriptional function (Fig. 4). While the EP2-mediated cAMP-dependent pathway may regulate Nurr1 function by upregulating its expression70,71, it is also possible that Nurr1’s protein stability and/or transcriptional activity may be regulated by altered levels of intracellular cAMP. Further studies are warranted for clarification of these mechanisms as well as for therapeutic development of these Nurr1 ligands.
In conclusion, in vitro and in vivo studies have recently provided strong evidence that Nurr1-activating compounds/ligands have the potential to protect dopaminergic neurons in various PD-related cell and animal models. Consequently, modulators that enhance Nurr1 function have great potential to be developed as novel mechanism-based disease-modifying therapeutics for PD from a transcriptional regulation perspective.
References
Mangelsdorf, D. J. et al. The nuclear receptor superfamily: the second decade. Cell 83, 835–839 (1995).
Kliewer, S. A., Lehmann, J. M. & Willson, T. M. Orphan nuclear receptors: shifting endocrinology into reverse. Science 284, 757–760 (1999).
Pawlak, A., Strzadala, L. & Kalas, W. Non-genomic effects of the NR4A1/Nur77/TR3/NGFIB orphan nuclear receptor. Steroids 95, 1–6 (2015).
Zhang, L. et al. The Orphan Nuclear Receptor 4A1: a potential new therapeutic target for metabolic diseases. J. Diabetes Res. 2018, 9363461 (2018).
Decressac, M., Volakakis, N., Bjorklund, A. & Perlmann, T. NURR1 in Parkinson disease-from pathogenesis to therapeutic potential. Nat. Rev. Neurol. 9, 629–636 (2013).
Dong, J., Li, S., Mo, J. L., Cai, H. B. & Le, W. D. Nurr1-based therapies for Parkinson’s disease. CNS Neurosci. Ther. 22, 351–359 (2016).
Wang, Z. et al. Structure and function of Nurr1 identifies a class of ligand-independent nuclear receptors. Nature 423, 555–560 (2003).
de Vera, I. M. S. et al. Defining a canonical ligand-binding pocket in the orphan nuclear receptor Nurr1. Structure 27, 66–77 e65 (2019).
Bruning, J. M. et al. Covalent modification and regulation of the nuclear receptor Nurr1 by a dopamine metabolite. cell. Chem. Biol. 26, 674–685 e676 (2019).
Rajan, S. et al. PGE1 and PGA1 bind to Nurr1rroo and activate its transcriptional function. Nat. Chem. Biol. 16, 876–886 (2020).
Hegarty, S. V., Sullivan, A. M. & O’Keeffe, G. W. Midbrain dopaminergic neurons: a review of the molecular circuitry that regulates their development. Dev. Biol. 379, 123–138 (2013).
Jang, Y. & Jung, J. H. Direct conversion from skin fibroblasts to functional dopaminergic neurons for biomedical application. Biomed. Dermatol. 1, 4 (2017).
Alavian, K. N. et al. The lifelong maintenance of mesencephalic dopaminergic neurons by Nurr1 and engrailed. J. Biomed. Sci. 21, 27 (2014).
Maira, M., Martens, C., Philips, A. & Drouin, J. Heterodimerization between members of the Nur subfamily of orphan nuclear receptors as a novel mechanism for gene activation. Mol. Cell Biol. 19, 7549–7557 (1999).
Kim, K. S. Toward neuroprotective treatments of Parkinson’s disease. Proc. Natl Acad. Sci. USA 114, 3795–3797 (2017).
Zetterstrom, R. H. et al. Dopamine neuron agenesis in Nurr1-deficient mice. Science 276, 248–250 (1997).
Kadkhodaei, B. et al. Transcription factor Nurr1 maintains fiber integrity and nuclear-encoded mitochondrial gene expression in dopamine neurons. Proc. Natl Acad. Sci. USA 110, 2360–2365 (2013).
Volakakis, N. et al. NR4A orphan nuclear receptors as mediators of CREB-dependent neuroprotection. Proc. Natl Acad. Sci. USA 107, 12317–12322 (2010).
Saijo, K. et al. A Nurr1/CoREST pathway in microglia and astrocytes protects dopaminergic neurons from inflammation-induced death. Cell 137, 47–59 (2009).
Lallier, S. W., Graf, A. E., Waidyarante, G. R. & Rogers, L. K. Nurr1 expression is modified by inflammation in microglia. Neuroreport 27, 1120–1127 (2016).
Chen, X. X. et al. Nurr1 promotes neurogenesis of dopaminergic neuron and represses inflammatory factors in the transwell coculture system of neural stem cells and microglia. CNS Neurosci. Ther. 24, 790–800 (2018).
Decressac, M. et al. alpha-Synuclein-induced down-regulation of Nurr1 disrupts GDNF signaling in nigral dopamine neurons. Sci. Transl. Med. 4, 163ra156 (2012).
Le, W., Conneely, O. M., He, Y., Jankovic, J. & Appel, S. H. Reduced Nurr1 expression increases the vulnerability of mesencephalic dopamine neurons to MPTP-induced injury. J. Neurochem. 73, 2218–2221 (1999).
Chu, Y., Kompoliti, K., Cochran, E. J., Mufson, E. J. & Kordower, J. H. Age-related decreases in Nurr1 immunoreactivity in the human substantia nigra. J. Comp. Neurol. 450, 203–214 (2002).
Ordentlich, P., Yan, Y., Zhou, S. & Heyman, R. A. Identification of the antineoplastic agent 6-mercaptopurine as an activator of the orphan nuclear hormone receptor Nurr1. J. Biol. Chem. 278, 24791–24799 (2003).
Estlin, E. J. Continuing therapy for childhood acute lymphoblastic leukaemia: clinical and cellular pharmacology of methotrexate, 6-mercaptopurine and 6-thioguanine. Cancer Treat. Rev. 27, 351–363 (2001).
Present, D. H. 6-Mercaptopurine and other immunosuppressive agents in the treatment of Crohn’s disease and ulcerative colitis. Gastroenterol. Clin. North Am. 18, 57–71 (1989).
Hintermann, S., Hengerer, B. & Schmidt, B. Heterocyclic Compounds Useful as Nurr-1 Activators Vol. WO2004072050A1 (Novartis Ag, N.P.G., French, 2004).
Dubois, C., Hengerer, B. & Mattes, H. Identification of a potent agonist of the orphan nuclear receptor Nurr1. ChemMedChem 1, 955–958 (2006).
Garcia, A. A., Lardenois, P., Olivier, A. 2-Aryl-6-phenylimidazo[1,2-α]pyridine Derivatives, Preparation Thereof and Therapeutic Use Thereof. Vol. WO2008034974A1 (Sanofi-Aventis, 2008).
Smith, G. A. et al. A Nurr1 agonist causes neuroprotection in a Parkinson’s disease lesion model primed with the toll-like receptor 3 dsRNA inflammatory stimulant poly(I:C). PLoS ONE 10, e0121072 (2015).
Lesuisse, D. et al. Development of a novel NURR1/NOT agonist from hit to lead and candidate for the potential treatment of Parkinson’s disease. Bioorg. Med. Chem. Lett. 29, 929–932 (2019).
Kim, C. H. et al. Nuclear receptor Nurr1 agonists enhance its dual functions and improve behavioral deficits in an animal model of Parkinson’s disease. Proc. Natl Acad. Sci. USA 112, 8756–8761 (2015).
Park, T. Y. et al. Chloroquine modulates inflammatory autoimmune responses through Nurr1 in autoimmune diseases. Sci. Rep. 9, 15559 (2019).
Moxon, C. A., Gibbins, M. P., McGuinness, D., Milner, D. A. Jr. & Marti, M. New Insights into Malaria Pathogenesis. Annu Rev. Pathol. 15, 315–343 (2020).
Pinheiro, L. C. S., Feitosa, L. M., Gandi, M. O., Silveira, F. F. & Boechat, N. The development of novel compounds against malaria: quinolines, triazolpyridines, pyrazolopyridines and pyrazolopyrimidines. Molecules 24, 4095 (2019).
Hedya, S. A., Safar, M. M. & Bahgat, A. K. Hydroxychloroquine antiparkinsonian potential: Nurr1 modulation versus autophagy inhibition. Behav. Brain Res. 365, 82–88 (2019).
Moon, M. et al. Nurr1 (NR4A2) regulates Alzheimer’s disease-related pathogenesis and cognitive function in the 5XFAD mouse model. Aging Cell 18, e12866 (2019).
Moon, M. et al. Correlation between orphan nuclear receptor Nurr1 expression and amyloid deposition in 5XFAD mice, an animal model of Alzheimer’s disease. J. Neurochem. 132, 254–262 (2015).
Moon, H., et al. Pharmacological stimulation of Nurr1 promotes cell cycle progression in adult hippocampal neural stem cells. Int. J. Mol. Sci. 21, 4 (2019).
Kinoshita, K. et al. A Nurr1 agonist amodiaquine attenuates inflammatory events and neurological deficits in a mouse model of intracerebral hemorrhage. J. Neuroimmunol. 330, 48–54 (2019).
Schrezenmeier, E. & Dorner, T. Mechanisms of action of hydroxychloroquine and chloroquine: implications for rheumatology. Nat. Rev. Rheumatol. 16, 155–166 (2020).
Floris, A. et al. Protective Effects of Hydroxychloroquine against Accelerated Atherosclerosis in Systemic Lupus Erythematosus. Mediators Inflamm. 2018, 3424136 (2018).
Danza, A., Grana, D., Goni, M., Vargas, A. & Ruiz-Irastorza, G. [Hydroxychloroquine for autoimmune diseases]. Rev. Med. Chil. 144, 232–240 (2016).
Sekiya, T. et al. The nuclear orphan receptor Nr4a2 induces Foxp3 and regulates differentiation of CD4+ T cells. Nat. Commun. 2, 269 (2011).
Hamers, A. A., Hanna, R. N., Nowyhed, H., Hedrick, C. C. & de Vries, C. J. NR4A nuclear receptors in immunity and atherosclerosis. Curr. Opin. Lipido. 24, 381–385 (2013).
Won, H. Y. & Hwang, E. S. Transcriptional modulation of regulatory T cell development by novel regulators NR4As. Arch. Pharm. Res. 39, 1530–1536 (2016).
Muramatsu, S. The current status of gene therapy for Parkinson’s disease. Ann. Neurosci. 17, 92–95 (2010).
Blits, B. & Petry, H. Perspective on the road toward gene therapy for Parkinson’s disease. Front. Neuroanat. 10, 128 (2016).
Yulug, B., Hanoglu, L. & Kilic, E. The neuroprotective role of vesicular monoamine transporter 2 in neurodegenerative diseases. Med. Chem. 11, 104–108 (2015).
Jayaramayya, K. et al. Unraveling correlative roles of dopamine transporter (DAT) and Parkin in Parkinson’s disease (PD)—a road to discovery? Brain Res. Bull. 157, 169–179 (2020).
Bisaglia, M., Mammi, S. & Bubacco, L. Kinetic and structural analysis of the early oxidation products of dopamine: analysis of the interactions with alpha-synuclein. J. Biol. Chem. 282, 15597–15605 (2007).
Pham, C. L. et al. Dopamine and the dopamine oxidation product 5,6-dihydroxylindole promote distinct on-pathway and off-pathway aggregation of alpha-synuclein in a pH-dependent manner. J. Mol. Biol. 387, 771–785 (2009).
Jang, Y., Kim, M. & Hwang, S. W. Molecular mechanisms underlying the actions of arachidonic acid-derived prostaglandins on peripheral nociception. J. Neuroinflammation 17, 30 (2020).
Hansen, H. S. Dietary essential fatty acids and in vivo prostaglandin production in mammals. World Rev. Nutr. Diet. 42, 102–134 (1983).
Vinayavekhin, N. & Saghatelian, A. Discovery of a protein-metabolite interaction between unsaturated fatty acids and the nuclear receptor Nur77 using a metabolomics approach. J. Am. Chem. Soc. 133, 17168–17171 (2011).
de Vera, I. M. et al. Identification of a binding site for unsaturated fatty acids in the orphan nuclear receptor Nurr1. ACS Chem. Biol. 11, 1795–1799 (2016).
Kagaya, S. et al. Prostaglandin A2 acts as a transactivator for NOR1 (NR4A3) within the nuclear receptor superfamily. Biol. Pharm. Bull. 28, 1603–1607 (2005).
Lakshmi, S. P., Reddy, A. T., Banno, A. & Reddy, R. C. Molecular, chemical, and structural characterization of prostaglandin A2 as a novel agonist for Nur77. Biochem. J. 476, 2757–2767 (2019).
Andersen, N. H. Dehydration of prostaglandins: study by spectroscopic method. J. Lipid Res. 10, 320–325 (1969).
Teagarden, D. L., Anderson, B. D. & Petre, W. J. Dehydration kinetics of prostaglandin E1 in a lipid emulsion. Pharm. Res. 6, 210–215 (1989).
Hubich, A. I. & Sholukh, M. V. Biochemistry of prostaglandins A. Biochemistry (Moscow) 71, 229–238 (2006).
Colville-Nash, P. R. & Gilroy, D. W. COX-2 and the cyclopentenone prostaglandins - a new chapter in the book of inflammation? Prostaglandins Other Lipid Mediat. 62, 33–43 (2000).
Karpisheh, V. et al. Prostaglandin E2 as a potent therapeutic target for treatment of colon cancer. Prostaglandins Other Lipid Mediat. 144, 106338 (2019).
Jang, Y. et al. Ghrelin receptor is activated by naringin and naringenin, constituents of a prokinetic agent Poncirus fructus. J. Ethnopharmacol. 148, 459–465 (2013).
Shim, W. S. et al. An aqueous extract of Poncirus fructus activates the prokinetic activity of 5-HT receptor subtype 4 without hERG interaction. J. Ethnopharmacol. 132, 328–333 (2010).
Jang, Y., Kim, T. K. & Shim, W. S. Naringin exhibits in vivo prokinetic activity via activation of ghrelin receptor in gastrointestinal motility dysfunction rats. Pharmacology 92, 191–197 (2013).
Carrasco, E., Casper, D. & Werner, P. PGE(2) receptor EP1 renders dopaminergic neurons selectively vulnerable to low-level oxidative stress and direct PGE(2) neurotoxicity. J. Neurosci. Res. 85, 3109–3117 (2007).
Carrasco, E., Werner, P. & Casper, D. Prostaglandin receptor EP2 protects dopaminergic neurons against 6-OHDA-mediated low oxidative stress. Neurosci. Lett. 441, 44–49 (2008).
Holla, V. R., Mann, J. R., Shi, Q. & DuBois, R. N. Prostaglandin E2 regulates the nuclear receptor NR4A2 in colorectal cancer. J. Biol. Chem. 281, 2676–2682 (2006).
McEvoy, A. N. et al. Activation of nuclear orphan receptor NURR1 transcription by NF-kappa B and cyclic adenosine 5’-monophosphate response element-binding protein in rheumatoid arthritis synovial tissue. J. Immunol. 168, 2979–2987 (2002).
Acknowledgements
This work was supported by the research fund of Hanyang University (HY-2018, 201800000003221).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
K.-S.K. is a co-founder of NurrON Pharmaceuticals. The remaining authors have no financial conflicts to disclose.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jang, Y., Kim, W., Leblanc, P. et al. Potent synthetic and endogenous ligands for the adopted orphan nuclear receptor Nurr1. Exp Mol Med 53, 19–29 (2021). https://doi.org/10.1038/s12276-021-00555-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s12276-021-00555-5