Abstract
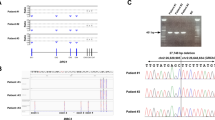
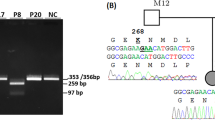
Primary ciliary dyskinesia (PCD) is a genetic disorder characterized by ciliary structural abnormalities and dysfunction, leading to chronic rhinosinusitis, otitis media with effusion, bronchiectasis, and infertility. Approximately half of Japanese PCD cases are attributed to variants in the dynein regulatory complex subunit 1 (DRC1) gene, predominantly featuring homogeneous deletions of exons 1–4 spanning 27,748 base pairs on chromosome 2. Here, we report 10 new PCD cases (9 families) in addition to 29 previously reported cases (24 families) caused by DRC1 variants. Among these 39 cases, biallelic DRC1 exon 1–4 deletions were detected in 38 (97.4%). These DRC1 deletions exhibited an identical breakpoint in all PCD cases in the Japanese and Korean populations, strongly suggesting a founder effect. In this study, we performed haplotype analysis, using a whole-exome sequencing dataset of 18 Japanese PCD patients harboring large biallelic DRC1 deletions. We estimated that the founder allele likely emerged 115.1 generations ago (95% confidence interval: 33.7–205.1), suggesting an origin of approximately 3050 years ago, coinciding with the transition from the Jomon period to the early Yayoi period in Japan. Considering the formation of the modern Japanese population, the founder with the DRC1 exon 1–4 deletion likely lived on the Korean peninsula, with the allele later transmitted to Japan through migration. This study provides insights into the origin of the DRC1 copy number variant, the most frequent PCD variant in the Japanese and Korean populations, highlighting the importance of understanding population-specific genetic variations in the context of human migration and disease prevalence.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Data generated in the course of this study have not been made publicly available in order to safeguard patient privacy and confidentiality. Investigators interested in accessing the data are encouraged to contact the corresponding author directly. Upon request, only anonymized data will be provided, contingent upon the approval of the study investigators and in compliance with all applicable privacy regulations and ethical guidelines.
References
Keicho N, Hijikata M, Miyabayashi A, Wakabayashi K, Yamada H, Ito M, et al. Impact of primary ciliary dyskinesia: beyond sinobronchial syndrome in Japan. Respir Investig. 2024;62:179–86.
Legendre M, Zaragosi LE, Mitchison HM. Motile cilia and airway disease. Semin Cell Dev Biol. 2021;110:19–33.
Xu Y, Feng G, Yano T, Masuda S, Nagao M, Gotoh S, et al. Characteristic genetic spectrum of primary ciliary dyskinesia in Japanese patients and global ethnic heterogeneity: population-based genomic variation database analysis. J Hum Genet. 2023;68:455–61.
Zariwala MA, Knowles MR, Leigh MW. Primary Ciliary Dyskinesia. In: Adam MP, Feldman J, Mirzaa GM, Pagon RA, Wallace SE, Bean LJH, et al., editors. GeneReviews((R)). Seattle (WA) 1993.
Takeuchi K, Xu Y, Kitano M, Chiyonobu K, Abo M, Ikegami K, et al. Copy number variation in DRC1 is the major cause of primary ciliary dyskinesia in the Japanese population. Mol Genet Genom Med. 2020;8:e1137.
Keicho N, Hijikata M, Morimoto K, Homma S, Taguchi Y, Azuma A, et al. Primary ciliary dyskinesia caused by a large homozygous deletion including exons 1-4 of DRC1 in Japanese patients with recurrent sinopulmonary infection. Mol Genet Genom Med. 2020;8:e1033.
Morimoto K, Hijikata M, Zariwala MA, Nykamp K, Inaba A, Guo TC, et al. Recurring large deletion in DRC1 (CCDC164) identified as causing primary ciliary dyskinesia in two Asian patients. Mol Genet Genom Med. 2019;7:e838.
Kim MJ, Kim S, Chae SW, Lee S, Yoon JG, Kim B, et al. Prevalence and founder effect of DRC1 exon 1–4 deletion in Korean patients with primary ciliary dyskinesia. J Hum Genet. 2023;68:369–74.
Kobayashi K, Nakahori Y, Mizuno K, Miyake M, Kumagai T, Honma A, et al. Founder-haplotype analysis in Fukuyama-type congenital muscular dystrophy (FCMD). Hum Genet. 1998;103:323–7.
Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature. 2015;526:68–74.
Takeuchi K, Abo M, Date H, Gotoh S, Kamijo A, Kaneko T, et al. Practical guide for the diagnosis and management of primary ciliary dyskinesia. Auris Nasus Larynx. 2024;51:553–68.
Takeuchi K, Kitano M, Kiyotoshi H, Ikegami K, Ogawa S, Ikejiri M, et al. A targeted next-generation sequencing panel reveals novel mutations in Japanese patients with primary ciliary dyskinesia. Auris Nasus Larynx. 2018;45:585–91.
Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i90.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60.
Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. https://doi.org/10.48550/arXiv.1207.3907.
Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, et al. Twelve years of SAMtools and BCFtools. Gigascience. 2021;10:giab008.
Tan A, Abecasis GR, Kang HM. Unified representation of genetic variants. Bioinformatics. 2015;31:2202–4.
Gandolfo LC, Bahlo M, Speed TP. Dating rare mutations from small samples with dense marker data. Genetics. 2014;197:1315–27.
Moorjani P, Sankararaman S, Fu Q, Przeworski M, Patterson N, Reich D. A genetic method for dating ancient genomes provides a direct estimate of human generation interval in the last 45,000 years. Proc Natl Acad Sci USA. 2016;113:5652–7.
Jombart T. Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24:1403–5.
Knaus BJ, Grunwald NJ. Vcfr: a package to manipulate and visualize variant call format data in R. Mol Ecol Resour. 2017;17:44–53.
Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat Genet. 2012;44:821–4.
Money D, Gardner K, Migicovsky Z, Schwaninger H, Zhong GY, Myles S. LinkImpute: fast and accurate genotype imputation for nonmodel organisms. G3 Genes. 2015;5(11):2383–90.
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–5.
Robinson JT, Thorvaldsdottir H, Turner D, Mesirov JP. igv.js: an embeddable JavaScript implementation of the Integrative Genomics Viewer (IGV). Bioinformatics. 2023;39:btac830.
Fassad MR, Patel MP, Shoemark A, Cullup T, Hayward J, Dixon M, et al. Clinical utility of NGS diagnosis and disease stratification in a multiethnic primary ciliary dyskinesia cohort. J Med Genet. 2020;57:322–30.
Hudson MJ, Nakagome S, Whitman JB. The evolving Japanese: the dual structure hypothesis at 30. Evol Hum Sci. 2020;2:e6.
Cooke NP, Mattiangeli V, Cassidy LM, Okazaki K, Kasai K, Bradley DG, et al. Genomic insights into a tripartite ancestry in the Southern Ryukyu Islands. Evol Hum Sci. 2023;5:e23.
Cooke NP, Mattiangeli V, Cassidy LM, Okazaki K, Stokes CA, Onbe S, et al. Ancient genomics reveals tripartite origins of Japanese populations. Sci Adv. 2021;7:eabh2419.
Ogami-Takamura K, Saiki K, Nishi K, Wakebe T, Endo D, Murai K, et al. Significant asymmetry of the bilateral upper extremities of a skeleton excavated from the Mashiki-Azamabaru Site, Okinawa Island, Japan. Biomed Res Int. 2021;2021:4884760.
Jinam T, Kawai Y, Kamatani Y, Sonoda S, Makisumi K, Sameshima H, et al. Genome-wide SNP data of Izumo and Makurazaki populations support inner-dual structure model for origin of Yamato people. J Hum Genet. 2021;66:681–87.
Watanabe Y, Naka I, Khor SS, Sawai H, Hitomi Y, Tokunaga K, et al. Analysis of whole Y-chromosome sequences reveals the Japanese population history in the Jomon period. Sci Rep. 2019;9:8556.
Hagihara Y, Nara T. Morphological features of the fibula in Jomon hunter-gatherers from the shell mounds of the Pacific coastal area. Am J Phys Anthropol. 2016;160:708–18.
Ito M, Morimoto K, Ohfuji T, Miyabayashi A, Wakabayashi K, Yamada H, et al. FOXJ1 variants causing primary ciliary dyskinesia with hydrocephalus: a case report from Japan. Intern Med. 2024;63:1433–7.
Xu Y, Ogawa S, Adachi Y, Sone N, Gotoh S, Ikejiri M, et al. A pediatric case of primary ciliary dyskinesia caused by novel copy number variation in PIH1D3. Auris Nasus Larynx. 2022;49:893–7.
Wallmeier J, Frank D, Shoemark A, Nothe-Menchen T, Cindric S, Olbrich H, et al. De novo mutations in FOXJ1 result in a motile ciliopathy with hydrocephalus and randomization of left/right body asymmetry. Am J Hum Genet. 2019;105:1030–39.
Hijikata M, Morimoto K, Takekoshi D, Shimoda M, Wakabayashi K, Miyabayashi A, et al. Analysis of aberrant splicing events and gene expression outliers in primary ciliary dyskinesia. Am J Respir Cell Mol Biol. 2023;68:702–05.
Horani A, Gupta DK, Xu J, Xu H, Carmen Puga-Molina LD, Santi CM, et al. The effect of Dnaaf5 gene dosage on primary ciliary dyskinesia phenotypes. JCI Insight. 2023;8:e168836.
Pivirotto A, Peles N, Hey J. Allele age estimators designed for whole genome datasets show only a modest decrease in accuracy when applied to whole exome datasets. bioRxiv. 2024:2024.02.01.578465. https://doi.org/10.1101/2024.02.01.578465.
Acknowledgements
We are grateful to the patient with PCD and their family members, who made this research possible. We thank the Center for Molecular Biology and Genetics at Mie University for performing the whole-exome sequencing.
Funding
This research was funded by JSPS Grants-in-Aid for Scientific Research (Grant Numbers 16K11210, 19K09886, 22K09665, and 22H03077) from the Ministry of Education, Culture, Sports, Science and Technology of Japan, the Takeda Science Foundation, and the Japan Agency for Medical Research and Development (AMED, Grant Number JP19ek0109410).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors have no conflict of interest to declare.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hashizume, R., Xu, Y., Ikejiri, M. et al. A 3000-year-old founder variant in the DRC1 gene causes primary ciliary dyskinesia in Japan and Korea. J Hum Genet (2024). https://doi.org/10.1038/s10038-024-01289-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s10038-024-01289-8