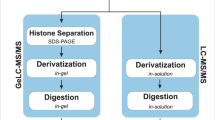
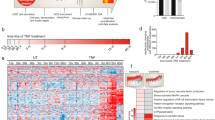
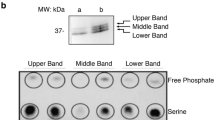
Abstract
Here we report an unbiased analysis of the cytotoxic T lymphocyte (CTL) serine-threonine phosphoproteome by high-resolution mass spectrometry. We identified approximately 2,000 phosphorylations in CTLs, of which approximately 450 were controlled by T cell antigen receptor (TCR) signaling. A significantly overrepresented group of molecules identified included transcription activators, corepressors and chromatin regulators. A focus on chromatin regulators showed that CTLs had high expression of the histone deacetylase HDAC7 but continually phosphorylated and exported this transcriptional repressor from the nucleus. Dephosphorylation of HDAC7 resulted in its accumulation in the nucleus and suppressed expression of genes encoding key cytokines, cytokine receptors and adhesion molecules that determine CTL function. Screening of the CTL phosphoproteome has thus identified intrinsic pathways of serine-threonine phosphorylation that target chromatin regulators and determine the CTL functional program.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Matthews, S.A. & Cantrell, D.A. New insights into the regulation and function of serine/threonine kinases in T lymphocytes. Immunol. Rev. 228, 241–252 (2009).
Lin, J.X., Spolski, R. & Leonard, W.J. Critical role for Rsk2 in T-lymphocyte activation. Blood 111, 525–533 (2008).
Salmond, R.J., Emery, J., Okkenhaug, K. & Zamoyska, R. MAPK, phosphatidylinositol 3-kinase, and mammalian target of rapamycin pathways converge at the level of ribosomal protein S6 phosphorylation to control metabolic signaling in CD8 T cells. J. Immunol. 183, 7388–7397 (2009).
Sinclair, L.V. et al. Phosphatidylinositol-3-OH kinase and nutrient-sensing mTOR pathways control T lymphocyte trafficking. Nat. Immunol. 9, 513–521 (2008).
Waugh, C., Sinclair, L., Finlay, D., Bayascas, J.R. & Cantrell, D. Phosphoinositide (3,4,5)-triphosphate binding to phosphoinositide-dependent kinase 1 regulates a protein kinase B/Akt signaling threshold that dictates T-cell migration, not proliferation. Mol. Cell. Biol. 29, 5952–5962 (2009).
Rao, R.R., Li, Q., Odunsi, K. & Shrikant, P.A. The mTOR kinase determines effector versus memory CD8+ T cell fate by regulating the expression of transcription factors T-bet and Eomesodermin. Immunity 32, 67–78 (2010).
Araki, K. et al. mTOR regulates memory CD8 T-cell differentiation. Nature 460, 108–112 (2009).
Pearce, E.L. et al. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature 460, 103–107 (2009).
Finlay, D.K. et al. Phosphoinositide-dependent kinase 1 controls migration and malignant transformation but not cell growth and proliferation in PTEN-null lymphocytes. J. Exp. Med. 206, 2441–2454 (2009).
Ong, S.E. et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 (2002).
Brockmeyer, C. et al. T cell receptor (TCR)-induced tyrosine phosphorylation dynamics identifies themis as a new TCR signalosome component. J. Biol. Chem. published online, doi:10.1074//jbc.M110.201236 (28 December 2010).
Mayya, V. et al. Quantitative phosphoproteomic analysis of T cell receptor signaling reveals system-wide modulation of protein-protein interactions. Sci. Signal. 2, ra46 (2009).
Nguyen, V. et al. A new approach for quantitative phosphoproteomic dissection of signaling pathways applied to T cell receptor activation. Mol. Cell. Proteomics 8, 2418–2431 (2009).
Cornish, G.H., Sinclair, L.V. & Cantrell, D.A. Differential regulation of T-cell growth by IL-2 and IL-15. Blood 108, 600–608 (2006).
Weninger, W., Crowley, M.A., Manjunath, N. & von Andrian, U.H. Migratory properties of naive, effector, and memory CD8+ T cells. J. Exp. Med. 194, 953–966 (2001).
Kalia, V. et al. Prolonged interleukin-2Rα expression on virus-specific CD8+ T cells favors terminal-effector differentiation in vivo. Immunity 32, 91–103 (2010).
Pipkin, M.E. et al. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity 32, 79–90 (2010).
Malek, T.R. & Castro, I. Interleukin-2 receptor signaling: at the interface between tolerance and immunity. Immunity 33, 153–165 (2010).
Muller, M.R. & Rao, A. NFAT, immunity and cancer: a transcription factor comes of age. Nat. Rev. Immunol. 10, 645–656 (2010).
Wabnitz, G.H., Nebl, G., Klemke, M., Schroder, A.J. & Samstag, Y. Phosphatidylinositol 3-kinase functions as a Ras effector in the signaling cascade that regulates dephosphorylation of the actin-remodeling protein cofilin after costimulation of untransformed human T lymphocytes. J. Immunol. 176, 1668–1674 (2006).
Marklund, U. et al. Serine 16 of oncoprotein 18 is a major cytosolic target for the Ca2+/calmodulin-dependent kinase-Gr. Eur. J. Biochem. 225, 53–60 (1994).
Wang, Z. et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138, 1019–1031 (2009).
Pflum, M.K., Tong, J.K., Lane, W.S. & Schreiber, S.L. Histone deacetylase 1 phosphorylation promotes enzymatic activity and complex formation. J. Biol. Chem. 276, 47733–47741 (2001).
Martin, M., Kettmann, R. & Dequiedt, F. Class IIa histone deacetylases: regulating the regulators. Oncogene 26, 5450–5467 (2007).
Dequiedt, F. et al. HDAC7, a thymus-specific class II histone deacetylase, regulates Nur77 transcription and TCR-mediated apoptosis. Immunity 18, 687–698 (2003).
Dequiedt, F. et al. Phosphorylation of histone deacetylase 7 by protein kinase D mediates T cell receptor-induced Nur77 expression and apoptosis. J. Exp. Med. 201, 793–804 (2005).
Parra, M., Kasler, H., McKinsey, T.A., Olson, E.N. & Verdin, E. Protein kinase D1 phosphorylates HDAC7 and induces its nuclear export after T-cell receptor activation. J. Biol. Chem. 280, 13762–13770 (2005).
Kao, H.Y. et al. Mechanism for nucleocytoplasmic shuttling of histone deacetylase 7. J. Biol. Chem. 276, 47496–47507 (2001).
Zhang, C.L. et al. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell 110, 479–488 (2002).
McKinsey, T.A., Zhang, C.L., Lu, J. & Olson, E.N. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 408, 106–111 (2000).
McKinsey, T.A., Zhang, C.L. & Olson, E.N. Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14–3-3 to histone deacetylase 5. Proc. Natl. Acad. Sci. USA 97, 14400–14405 (2000).
Matthews, S.A. et al. Essential role for protein kinase D family kinases in the regulation of class II histone deacetylases in B lymphocytes. Mol. Cell. Biol. 26, 1569–1577 (2006).
Martin, M., Kettmann, R. & Dequiedt, F. Class IIa histone deacetylases: conducting development and differentiation. Int. J. Dev. Biol. 53, 291–301 (2009).
Fischle, W. et al. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol. Cell 9, 45–57 (2002).
Li, X., Song, S., Liu, Y., Ko, S.H. & Kao, H.Y. Phosphorylation of the histone deacetylase 7 modulates its stability and association with 14–3-3 proteins. J. Biol. Chem. 279, 34201–34208 (2004).
Kasler, H.G. & Verdin, E. Histone deacetylase 7 functions as a key regulator of genes involved in both positive and negative selection of thymocytes. Mol. Cell. Biol. 27, 5184–5200 (2007).
Dubois, F. et al. Differential 14–3-3 affinity capture reveals new downstream targets of phosphatidylinositol 3-kinase signaling. Mol. Cell. Proteomics 8, 2487–2499 (2009).
Gao, J., Opiteck, G.J., Friedrichs, M.S., Dongre, A.R. & Hefta, S.A. Changes in the protein expression of yeast as a function of carbon source. J. Proteome Res. 2, 643–649 (2003).
Gao, C. et al. CRM1 mediates nuclear export of HDAC7 independently of HDAC7 phosphorylation and association with 14–3-3s. FEBS Lett. 580, 5096–5104 (2006).
Yang, X.J. & Seto, E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat. Rev. Mol. Cell Biol. 9, 206–218 (2008).
Parra, M., Mahmoudi, T. & Verdin, E. Myosin phosphatase dephosphorylates HDAC7, controls its nucleocytoplasmic shuttling, and inhibits apoptosis in thymocytes. Genes Dev. 21, 638–643 (2007).
Martin, M. et al. Protein phosphatase 2A controls the activity of histone deacetylase 7 during T cell apoptosis and angiogenesis. Proc. Natl. Acad. Sci. USA 105, 4727–4732 (2008).
Tamas, P. et al. LKB1 is essential for the proliferation of T-cell progenitors and mature peripheral T cells. Eur. J. Immunol. 40, 242–253 (2010).
Fox, C.J., Hammerman, P.S. & Thompson, C.B. The Pim kinases control rapamycin-resistant T cell survival and activation. J. Exp. Med. 201, 259–266 (2005).
Chang, S., Bezprozvannaya, S., Li, S. & Olson, E.N. An expression screen reveals modulators of class II histone deacetylase phosphorylation. Proc. Natl. Acad. Sci. USA 102, 8120–8125 (2005).
Seki, Y. et al. TIF1β regulates the pluripotency of embryonic stem cells in a phosphorylation-dependent manner. Proc. Natl. Acad. Sci. USA 107, 10926–10931 (2010).
Chang, C.W. et al. Phosphorylation at Ser473 regulates heterochromatin protein 1 binding and corepressor function of TIF1β/KAP1. BMC Mol. Biol. 9, 61 (2008).
McNulty, D.E. & Annan, R.S. Hydrophilic interaction chromatography reduces the complexity of the phosphoproteome and improves global phosphopeptide isolation and detection. Mol. Cell. Proteomics 7, 971–980 (2008).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008).
Tusher, V.G., Tibshirani, R. & Chu, G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98, 5116–5121 (2001).
Huang da, W., Sherman, B.T. & Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Acknowledgements
We thank members of the Biological Services Unit, R. Clarke of the Flow Cytometry Facility and members of the Cantrell laboratory for critical reading of the manuscript; and the Finnish DNA Microarray Centre at the Centre for Biotechnology (Turku, Finland) and N. Schurch for microarray analysis. Supported by the Wellcome Trust (065975/Z/01/A) and the Medical Research Council (J.G.).
Author information
Authors and Affiliations
Contributions
M.N.N., J.G. and C.F.-C. did the experiments and analyzed the results; N.M. supervised SILAC methodology and bioinformatic analysis; and M.N.N. and D.A.C. designed the experiments, analyzed the results and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4, Supplementary Tables 1–3 and Supplementary Methods (PDF 10880 kb)
Rights and permissions
About this article
Cite this article
Navarro, M., Goebel, J., Feijoo-Carnero, C. et al. Phosphoproteomic analysis reveals an intrinsic pathway for the regulation of histone deacetylase 7 that controls the function of cytotoxic T lymphocytes. Nat Immunol 12, 352–361 (2011). https://doi.org/10.1038/ni.2008
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.2008
This article is cited by
-
Regulation of histone deacetylase activities and functions by phosphorylation and its physiological relevance
Cellular and Molecular Life Sciences (2021)
-
Signals for antigen-independent differentiation of memory CD8+ T cells
Cellular and Molecular Life Sciences (2021)
-
Genetics of primary sclerosing cholangitis and pathophysiological implications
Nature Reviews Gastroenterology & Hepatology (2017)
-
The cytotoxic T cell proteome and its shaping by the kinase mTOR
Nature Immunology (2016)
-
Label-free quantitative phosphorylation analysis of human transgelin2 in Jurkat T cells reveals distinct phosphorylation patterns under PKA and PKC activation conditions
Proteome Science (2015)