Abstract
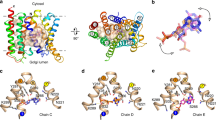
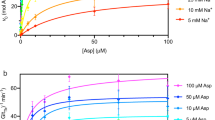
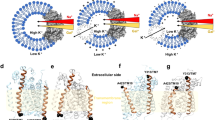
Transporter-related nutrient sensors, called transceptors, mediate nutrient activation of signaling pathways through the plasma membrane. The mechanism of action of transporting and nontransporting transceptors is unknown. We have screened 319 amino acid analogs to identify compounds that act on Gap1, a transporting amino acid transceptor in yeast that triggers activation of the protein kinase A pathway. We identified competitive and noncompetitive inhibitors of transport, either with or without agonist action for signaling, including nontransported agonists. Using substituted cysteine accessibility method (SCAM) analysis, we identified Ser388 and Val389 as being exposed into the amino acid binding site, and we show that agonist action for signaling uses the same binding site as used for transport. Our results provide the first insight, to our knowledge, into the mechanism of action of transceptors. They indicate that signaling requires a ligand-induced specific conformational change that may be part of but does not require the complete transport cycle.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Ozcan, S., Dover, J. & Johnston, M. Glucose sensing and signaling by two glucose receptors in the yeast Saccharomyces cerevisiae. EMBO J. 17, 2566–2573 (1998).
Didion, T., Regenberg, B., Jorgensen, M.U., Kielland-Brandt, M.C. & Andersen, H.A. The permease homologue Ssy1p controls the expression of amino acid and peptide transporter genes in Saccharomyces cerevisiae. Mol. Microbiol. 27, 643–650 (1998).
Iraqui, I. et al. Amino acid signaling in Saccharomyces cerevisiae: a permease-like sensor of external amino acids and F-Box protein Grr1p are required for transcriptional induction of the AGP1 gene, which encodes a broad-specificity amino acid permease. Mol. Cell. Biol. 19, 989–1001 (1999).
Klasson, H., Fink, G.R. & Ljungdahl, P.O. Ssy1p and Ptr3p are plasma membrane components of a yeast system that senses extracellular amino acids. Mol. Cell. Biol. 19, 5405–5416 (1999).
Holsbeeks, I., Lagatie, O., Van Nuland, A., Van de Velde, S. & Thevelein, J.M. The eukaryotic plasma membrane as a nutrient-sensing device. Trends Biochem. Sci. 29, 556–564 (2004).
Biswas, K. & Morschhauser, J. The Mep2p ammonium permease controls nitrogen starvation-induced filamentous growth in Candida albicans. Mol. Microbiol. 56, 649–669 (2005).
Lorenz, M.C. & Heitman, J. The MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae. EMBO J. 17, 1236–1247 (1998).
Hyde, R., Cwiklinski, E.L., MacAulay, K., Taylor, P.M. & Hundal, H.S. Distinct sensor pathways in the hierarchical control of SNAT2, a putative amino acid transceptor, by amino acid availability. J. Biol. Chem. 282, 19788–19798 (2007).
Goberdhan, D.C., Meredith, D., Boyd, C.A. & Wilson, C. PAT-related amino acid transporters regulate growth via a novel mechanism that does not require bulk transport of amino acids. Development 132, 2365–2375 (2005).
Hyde, R., Taylor, P.M. & Hundal, H.S. Amino acid transporters: roles in amino acid sensing and signalling in animal cells. Biochem. J. 373, 1–18 (2003).
Lalonde, S. et al. The dual function of sugar carriers. Transport and sugar sensing. Plant Cell 11, 707–726 (1999).
Thevelein, J.M. & de Winde, J.H. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 33, 904–918 (1999).
Thevelein, J.M. et al. Nutrient sensing systems for rapid activation of the protein kinase A pathway in yeast. Biochem. Soc. Trans. 33, 253–256 (2005).
Donaton, M.C. et al. The Gap1 general amino acid permease acts as an amino acid sensor for activation of protein kinase A targets in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 50, 911–929 (2003).
Van Nuland, A. et al. Ammonium permease-based sensing mechanism for rapid ammonium activation of the protein kinase A pathway in yeast. Mol. Microbiol. 59, 1485–1505 (2006).
Giots, F., Donaton, M.C. & Thevelein, J.M. Inorganic phosphate is sensed by specific phosphate carriers and acts in concert with glucose as a nutrient signal for activation of the protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 47, 1163–1181 (2003).
Grenson, M. Inactivation-reactivation process and repression of permease formation regulate several ammonia-sensitive permeases in the yeast Saccharomyces cerevisiae. Eur. J. Biochem. 133, 135–139 (1983).
Hein, C. & Andre, B. A C-terminal di-leucine motif and nearby sequences are required for NH4(+)-induced inactivation and degradation of the general amino acid permease, Gap1p, of Saccharomyces cerevisiae. Mol. Microbiol. 24, 607–616 (1997).
Stanbrough, M. & Magasanik, B. Transcriptional and posttranslational regulation of the general amino acid permease of Saccharomyces cerevisiae. J. Bacteriol. 177, 94–102 (1995).
Andre, B. An overview of membrane transport proteins in Saccharomyces cerevisiae. Yeast 11, 1575–1611 (1995).
Wipf, D. et al. Conservation of amino acid transporters in fungi, plants and animals. Trends Biochem. Sci. 27, 139–147 (2002).
Gilstring, C.F. & Ljungdahl, P.O. A method for determining the in vivo topology of yeast polytopic membrane proteins demonstrates that Gap1p fully integrates into the membrane independently of Shr3p. J. Biol. Chem. 275, 31488–31495 (2000).
Hu, L.A. & King, S.C. Functional sensitivity of polar surfaces on transmembrane helix 8 and cytoplasmic loop 8–9 of the Escherichia coli GABA (4-aminobutyrate) transporter encoded by gabP: mutagenic analysis of a consensus amphipathic region found in transporters from bacteria to mammals. Biochem. J. 330, 771–776 (1998).
King, S.C., Hu, L.A. & Pugh, A. Induction of substrate specificity shifts by placement of alanine insertions within the consensus amphipathic region of the Escherichia coli GABA (gamma-aminobutyric acid) transporter encoded by gabP. Biochem. J. 376, 645–653 (2003).
Wu, B. et al. Competitive intra- and extracellular nutrient sensing by the transporter homologue Ssy1p. J. Cell Biol. 173, 327–331 (2006).
App, H. & Holzer, H. Purification and characterization of neutral trehalase from the yeast ABYS1 mutant. J. Biol. Chem. 264, 17583–17588 (1989).
Durnez, P. et al. Activation of trehalase during growth induction by nitrogen sources in the yeast Saccharomyces cerevisiae depends on the free catalytic subunits of cAMP-dependent protein kinase, but not on functional Ras proteins. Yeast 10, 1049–1064 (1994).
Hirimburegama, K. et al. Nutrient-induced activation of trehalase in nutrient-starved cells of the yeast Saccharomyces cerevisiae: cAMP is not involved as second messenger. J. Gen. Microbiol. 138, 2035–2043 (1992).
Uno, I., Matsumoto, K., Adachi, K. & Ishikawa, T. Genetic and biochemical evidence that trehalase is a substrate of cAMP-dependent protein kinase in yeast. J. Biol. Chem. 258, 10867–10872 (1983).
Ma, P., Wera, S., Van Dijck, P. & Thevelein, J.M. The PDE1-encoded low-affinity phosphodiesterase in the yeast Saccharomyces cerevisiae has a specific function in controlling agonist-induced cAMP signaling. Mol. Biol. Cell 10, 91–104 (1999).
Mbonyi, K., van Aelst, L., Arguelles, J.C., Jans, A.W. & Thevelein, J.M. Glucose-induced hyperaccumulation of cyclic AMP and defective glucose repression in yeast strains with reduced activity of cyclic AMP-dependent protein kinase. Mol. Cell. Biol. 10, 4518–4523 (1990).
De Craene, J.O., Soetens, O. & Andre, B. The Npr1 kinase controls biosynthetic and endocytic sorting of the yeast Gap1 permease. J. Biol. Chem. 276, 43939–43948 (2001).
Risinger, A.L., Cain, N.E., Chen, E.J. & Kaiser, C.A. Activity-dependent reversible inactivation of the general amino acid permease. Mol. Biol. Cell 17, 4411–4419 (2006).
Rubio-Texeira, M. & Kaiser, C.A. Amino acids regulate retrieval of the yeast general amino acid permease from the vacuolar targeting pathway. Mol. Biol. Cell 17, 3031–3050 (2006).
Springael, J.Y. & Andre, B. Nitrogen-regulated ubiquitination of the Gap1 permease of Saccharomyces cerevisiae. Mol. Biol. Cell 9, 1253–1263 (1998).
Grenson, M., Hou, C. & Crabeel, M. Multiplicity of the amino acid permeases in Saccharomyces cerevisiae. IV. Evidence for a general amino acid permease. J. Bacteriol. 103, 770–777 (1970).
Poulsen, P., Gaber, R.F. & Kielland-Brandt, M.C. Hyper- and hyporesponsive mutant forms of the Saccharomyces cerevisiae Ssy1 amino acid sensor. Mol. Membr. Biol. 25, 164–176 (2008).
Ozcan, S., Dover, J., Rosenwald, A.G., Wolfl, S. & Johnston, M. Two glucose transporters in Saccharomyces cerevisiae are glucose sensors that generate a signal for induction of gene expression. Proc. Natl. Acad. Sci. USA 93, 12428–12432 (1996).
Schwoppe, C., Winkler, H.H. & Neuhaus, H.E. Properties of the glucose-6-phosphate transporter from Chlamydia pneumoniae (HPTcp) and the glucose-6-phosphate sensor from Escherichia coli (UhpC). J. Bacteriol. 184, 2108–2115 (2002).
Schwoppe, C., Winkler, H.H. & Neuhaus, H.E. Connection of transport and sensing by UhpC, the sensor for external glucose-6-phosphate in Escherichia coli. Eur. J. Biochem. 270, 1450–1457 (2003).
Faham, S. et al. The crystal structure of a sodium galactose transporter reveals mechanistic insights into Na+/sugar symport. Science 321, 810–814 (2008).
Holyoake, J. & Sansom, M.S. Conformational change in an MFS protein: MD simulations of LacY. Structure 15, 873–884 (2007).
Yamashita, A., Singh, S.K., Kawate, T., Jin, Y. & Gouaux, E. Crystal structure of a bacterial homologue of Na+/Cl–dependent neurotransmitter transporters. Nature 437, 215–223 (2005).
Bechet, J., Greenson, M. & Wiame, J.M. Mutations affecting the repressibility of arginine biosynthetic enzymes in Saccharomyces cerevisiae. Eur. J. Biochem. 12, 31–39 (1970).
Jauniaux, J.C. & Grenson, M. GAP1, the general amino acid permease gene of Saccharomyces cerevisiae. Nucleotide sequence, protein similarity with the other bakers yeast amino acid permeases, and nitrogen catabolite repression. Eur. J. Biochem. 190, 39–44 (1990).
Bonneaud, N. et al. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors. Yeast 7, 609–615 (1991).
Tamas, M.J., Rep, M., Thevelein, J.M. & Hohmann, S. Stimulation of the yeast high osmolarity glycerol (HOG) pathway: evidence for a signal generated by a change in turgor rather than by water stress. FEBS Lett. 472, 159–165 (2000).
Acknowledgements
We thank W. Verheyden and R. Wicik for technical help with the experiments, N. Vangoethem for help with preparation of the figures and B. André (Université Libre de Bruxelles) for the kind gift of strains, plasmids and antibodies. This work was supported by a Return Grant from the Belgian Federal Science Policy Office to M.V. and by grants from the Fund for Scientific Research - Flanders, Interuniversity Attraction Poles Network P5/30 and P6/14 and the Research Fund of the Katholieke Universiteit Leuven (Concerted Research Actions).
Author information
Authors and Affiliations
Contributions
G.V.Z. and B.M.B. contributed mainly to the execution of the experimental work; M.V. and J.M.T. contributed to the design and discussion of the experimental work.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8, Supplementary Table 1 and Supplementary Discussion (PDF 475 kb)
Rights and permissions
About this article
Cite this article
Van Zeebroeck, G., Bonini, B., Versele, M. et al. Transport and signaling via the amino acid binding site of the yeast Gap1 amino acid transceptor. Nat Chem Biol 5, 45–52 (2009). https://doi.org/10.1038/nchembio.132
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.132
This article is cited by
-
Role of Amino Acid Metabolism in the Virulence of Human Pathogenic Fungi
Current Clinical Microbiology Reports (2019)
-
Metabolic engineering of arginine permeases to reduce the formation of urea in Saccharomyces cerevisiae
World Journal of Microbiology and Biotechnology (2018)
-
Identification and characterization of the glucose dual-affinity transport system in Neurospora crassa: pleiotropic roles in nutrient transport, signaling, and carbon catabolite repression
Biotechnology for Biofuels (2017)
-
Effects of three permeases on arginine utilization in Saccharomyces cerevisiae
Scientific Reports (2016)
-
Identifying novel protein phenotype annotations by hybridizing protein–protein interactions and protein sequence similarities
Molecular Genetics and Genomics (2016)