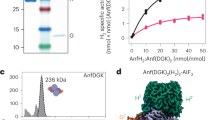
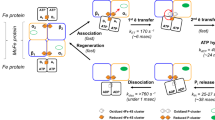
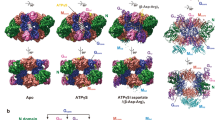
Abstract
The coupling of ATP hydrolysis to electron transfer by the enzyme nitrogenase during biological nitrogen fixation is an important example of a nucleotide-dependent transduction mechanism. The crystal structure has been determined for the complex between the Fe-protein and MoFe-protein components of nitrogenase stabilized by ADP·AIF4–, previously used as a nucleoside triphosphate analogue in nucleotide-switch proteins. The structure reveals that the dimeric Fe-protein has undergone substantial conformational changes. The β-phosphate and AIF4– groups are stabilized through intersubunit contacts that are critical for catalysis and the redox centre is repositioned to facilitate electron transfer. Interactions in the nitrogenase complex have broad implications for signal and energy transduction mechanisms in multiprotein complexes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Stiefel, E. I.,, Coucouvanis, D. & Newton, W. E. (eds) Molybdenum Enzymes, Cofactors and Model Systems (Am. Chem. Soc., Washington, DC, 1993).
Mortenson, L. E., Seefeldt, L. C., Morgan, T. V. & Bolin, J. T. The role of metal-clusters and MgATP in nitrogenase catalysis. Adv. Enzymol. Rel. Areas Mol. Biol. 67, 299–274 (1993).
Howard, J. B. & Rees, D. C. Nitrogenase: A nucleotide-dependent molecular switch. Annu. Rev. Biochem. 63, 235–264 (1994).
Peters, J. W., Fisher, K. & Dean, D. R. Nitrogenase structure and function: A biochemical-genetic perspective. Annu. Rev. Microbiol. 49, 335–366 (1995).
Burgess, B. K. & Lowe, D. J. Mechanism of molybdenum nitrogenase. Chem. Rev. 96, 2983–3011 (1996).
Howard, J. B. & Rees, D. C. Structural basis of biological nitrogen fixation. Chem. Rev. 96, 2965–2982 (1996).
Renner, K. A. & Howard, J. B. Aluminium fluoride inhibition of nitrogenase: stabilization of a nucleotide: Fe-protein : MoFe-protein complex. Biochemistry 35, 5353–5358 (1996).
Duyvis, M., Wassink, H. & Haaker, H. Formation and characterization of a transition state complex of Azotobacter vinelandii nitrogenase. FEBS Lett. 380, 233–236 (1996).
Fisher, A. J. et al. X-ray structures of the myosin motor domain of Dictyostelium discoideum complexed with MgADP·BeFx and MgADP·AlF−4 . Biochemistry 34, 8960–8972 (1995).
Sondek, J., Lambright, D. G., Noel, J. P., Hamm, H. E. & Sigler, P. B. GTPase mechanisms of G proteins from the 1.7-Å crystal structure of transducin α·GDP·ALF−4 . Nature 372, 276–279 (1994).
Coleman, D. E. et al. Structures of active conformers of Giα1 and the mechanism of GTP hydrolysis. Science 265, 1405–1412 (1994).
Mittal, R., Ahmadian, M. R., Goody, R. S. & Wittinghofer, A. Formation of a transition-state analog of the Ras GTPase reaction by Ras-GDP, tetrafluoroaluminate, and GTPase-activating proteins. Science 273, 115–117 (1996).
Kim, J. & Rees, D. C. Structural models for the metal centers in the nitrogenase molybdenum-iron protein. Science 257, 1677–1682 (1992).
Kim, J. & Rees, D. C. Crystallographic structure and functional implications of the nitrogenase molybdenum-iron protein from Azotobacter vinelandii. Nature 360, 553–560 (1992).
Georgiadis, M. M. et al. Crystallographic structure of the nitrogenase iron protein from Azotobacter vinelandii. Science 257, 1653–1659 (1992).
Bolin, J. T., Campobasso, N., Muchmore, S. W., Morgan, T. V. & Mortenson, L. E. in Molybdenum Enzymes, Cofactors and Model Systems. ACS Symposium Series No. 535 (eds Stiefel, E. I., Coucouvanis, D. & Newton, W. E.) 186–195 (Am. Chem. Soc., Washington, 1993).
Peters, J. P. et al. Redox-dependent structural changes in the nitrogenase P-cluster. Biochemistry 36, 1181–1187 (1997).
Willing, A. & Howard, J. B. Cross-linking site in Azotobacter vinelandii complex. J. Biol. Chem. 265, 6596–6599 (1990).
Howard, J. B. in Molybdenum Enzymes, Cofactors and Model Systems. ACS Symposium Series No. 535 (eds Stiefel, E. I., Coucouvanis, D. & Newton, W. E.) 271–289 (Am. Chem. Soc., Washington, 1993).
Ludden, P. W. & Roberts, G. P. Regulation of nitrogenase activity by reversible ADP ribosylation. Curr. Top. Cell. Regul. 30, 23–56 (1989).
Wolle, D., Kim, C., Dean, D. & Howard, J. B. Ionic interactions in the nitrogenase complex. J. Biol. Chem. 267, 3667–3673 (1992).
Chen, L. et al. MgATP-induced conformational changes in the iron protein from A. vinelandii as studied by small-angle x-ray scattering. J. Biol. Chem. 269, 3290–3294 (1994).
Pai, E. F. et al. Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 Å resolution—implications for the mechanism of GTP hydrolysis. EMBO J. 9, 2351–2359 (1990).
Noel, J. P., Hamm, H. E. & Sigler, P. B. The 2.2 Å crystal structure of transducin-α complexed with GTP-yS. Nature 366, 654–663 (1993).
Lanzilotta, W. N., Ryle, M. J. & Seefeldt, L. C. Nucleotide hydrolysis and protein conformational changes in Azotobacter vinelandii nitrogenase iron protein: defining the function of aspartate 129. Biochemistry 34, 10712–10723 (1995).
Ryle, M. J. & Seefeldt, L. C. Elucidation of a MgATP signal transduction pathway in the nitrogenase iron protein: formation of a conformation resembling the MgATP-bound state by protein engineering. Biochemistry 35, 4766–4775 (1996).
Seefeldt, I. C., Morgan, T. V., Dean, D. R. & Mortenson, L. E. Mapping the site(s) of MgATP and MgADP interaction with the nitrogenase of Azotobacter vinelandii. J. Biol. Chem. 267, 6680–6688 (1992).
Ryle, M. J., Lanzilotta, W. N., Mortenson, L. E., Watt, G. D. & Seefeldt, L. C. Evidence for a central role of lysine 15 of Azotobacter vinelandii nitrogenase iron protein in nucleotide binding and protein conformational changes. J. Biol. Chem. 270, 13112–13117 (1995).
Seefeldt, L. C. & Mortenson, L. E. Increasing nitrogenase catalytic efficiency for MgATP by changing serine 16 of its Fe protein to threonine: Use of Mn 2+ to show interaction of serine 16 with Mg2+. Prot. Sci. 2, 93–102 (1993).
Wolle, D., Dean, D. R. & Howard, J. B. Nucleotide-iron-sulfur cluster signal transduction in the nitrogenase iron-protein: the role of Asp125. Science 258, 992–995 (1992).
Frech, M. et al. Role of glutamine-61 in the hydrolysis of GTP by p21H-ras. An experimental and theoretical study. Biochemistry 33, 3237–3244 (1994).
Maegley, K. A., Admiraal, S. J. & Herschlag, D. Ras-catalyzed hydrolysis of GTP: A new perspective from model studies. Proc. Natl Acad. Sci. USA 93, 8160–8166 (1996).
Brownbridge, G. G., Lowe, P. N., Moore, K. J. M., Skinner, R. H. & Webb, M. R. Interaction of GTPase activating proteins (GAPs) with p21ras measured by a novel fluorescence anisotropy method: Essential role of Arg-903 of GAP in activation of GTP hydrolysis on p21ras. J. Biol. Chem. 268, 10914–10919 (1993).
Otwinowski, Z. in Data Collection and Processing (eds Sawyer, L., Issacs, N. & Bailey, S.) 56–62 (SERC Daresbury Laboratory, UK, 1993).
Bailey, S. The CCP4 suite—programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994).
Navaza, J. AMORE—an automated package for molecular replacement. Acta Crystallogr. A 50, 157–163 (1994).
Kleywegt, G. J. & Jones, T. A. in First Map to Final Model (eds Bailey, S., Hubbard, R. & Waller, D.) 59–66 (SERC Daresbury Laboratory, UK, 1994).
Jones, T. A., Zhou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Brünger, A. T. X-PLOR version 3.1—A system for X-ray crystallography and NMR (Yale University Press, New Haven and London, 1992).
Laskowski, R. A., McArthur, M. W., Moss, D. S. & Thornton, J. M. PROCHECK—a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993).
Kabsch, W. A solution for the best rotation to relate two sets of vectors. Acta Crystallogr. A 32, 922–923 (1976).
Kraulis, P. J. MOLSCRIPT—a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950 (1991).
Bacon, D. J. & Anderson, W. F. A fast algorithm for rendering space-filling molecule pictures. J. Mol. Graph. 6, 219–220 (1988).
Merritt, E. A. & Murphy, M. E. P. Raster3D Version 2.0-a program for photorealistic molecular graphics. Acta Crystallogr. D 50, 869–873 (1994).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Schindelin, H., Kisker, C., Schlessman, J. et al. Structure of ADP·AIF4–-stabilized nitrogenase complex and its implications for signal transduction. Nature 387, 370–376 (1997). https://doi.org/10.1038/387370a0
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/387370a0
This article is cited by
-
Fe protein docking transduces conformational changes to MoFe nitrogenase active site in a nucleotide-dependent manner
Communications Chemistry (2023)
-
Chainlike products from the reaction of aluminum nanoparticles in HF atmosphere: an atomic insight
Journal of Materials Science (2022)
-
The E. coli MinCDE system in the regulation of protein patterns and gradients
Cellular and Molecular Life Sciences (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.