Abstract
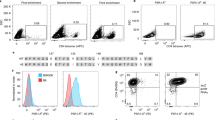
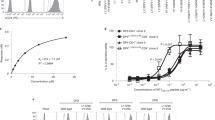
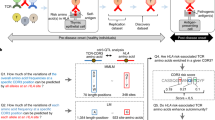
The serologically detectable molecules encoded by the H–2 complex, the major histocompatibility complex (MHC) of the mouse, fall into two classes—class I and class II (ref. 1). The class I molecules encoded by the K and D loci have a molecular weight of 44,000 and are noncovalently associated with β2 microglobulin which is not controlled by the MHC. The class II molecules are of two kinds, A and E, each consisting of two noncovalently associated polypeptide chains, α (Mr ∼34,000) and β (Mr ∼28,000)2,3. Three of the four chains, Aα, Aβ and Eβ, are controlled by loci in the I-A subregion, whereas the locus controlling the Eα chain is located in the I-A subregion of the H–2 complex. Thus the loci coding for the Eα and Eβ chains are separated by at least one (J) and perhaps more loci3,4. It has been shown that the Eα and Eβ chains are synthesized independently, and that the Eα chain is required for the insertion of the Eβ chain in the plasma membrane4. We demonstrate here that the EαEβ complex (the E molecule) can evoke in vitro cell-mediated lymphocytotoxicity (CML) without previous sensitization in vivo, and that the strong CML reactivity is directed against allele-specific determinants on the highly polymorphic Eβ chain.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Klein, J. Science 203, 516–521 (1979).
Cullen, S. E., Freed, J. H. & Nathenson, S. G. Transplant Rev. 30, 236–239 (1976).
Uhr, J. W., Capra, J. D., Vitetta, E. S. & Cook, R. G. Science 206, 292–297 (1979).
Jones, P. P., Murphy, D. B. & McDevitt, H. O. J. exp. Med. 148, 925–939 (1978).
Juretić, A. et al. Immunogenetics (submitted).
Lozner, E. C., Sachs, D. H., Shearer, G. M. & Terry, W. D. Science 183, 757–759 (1974).
David, C. S. & Shreffler, D. C. Transplantation 18, 313–321 (1974).
Klein, J., Flaherty, L., VandeBerg, J. L. & Shreffler, D. C. Immunogenetics 6, 489–512 (1978).
Egorov, J. K., Apt, A. S. & Vedernikov, A. A. Immunogenetics 5, 285–290 (1977).
Fischer Lindahl, K. & Hausmann, B. Immunogenetics (in the press).
Klein, J. et al. Nature (submitted).
Lafuse, W. P., McCormick, J. E. & David, C. S. J. exp. Med. 151, 709–715 (1980).
Benecerraf, B. & Germain, R. N. Immun. Rev. 38, 70–119 (1978).
Peck, B. A., Klein, J. & Wigzell, H. J. J. Immun. 125, 1078–1086 (1980).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
JuretiĆ, A., Nagy, Z. & Klein, J. Detection of CML determinants associated with H–2 controlled Eβ and Eαchains. Nature 289, 308–310 (1981). https://doi.org/10.1038/289308a0
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/289308a0
This article is cited by
-
Polymorphism of minor histocompatibility genes in wild mice
Immunogenetics (1983)
-
Cytotoxic monoclonal antibody specific for the Lyt-1.2 antigen
Immunogenetics (1982)
-
Clonal analysis of the specificity of alloreactive cells: ?Dominance? of E? reactive clones
Immunogenetics (1982)
-
Cytolytic T cells activated by H-2-controlled E molecules cross-react with A molecules
Immunogenetics (1982)
-
Serological cross-reactivity between products of separate I regions
Immunogenetics (1982)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.