Long-range sequence assembly in whole-genome shotgun sequencing
Keywords
Flag Inappropriate
Delete Content
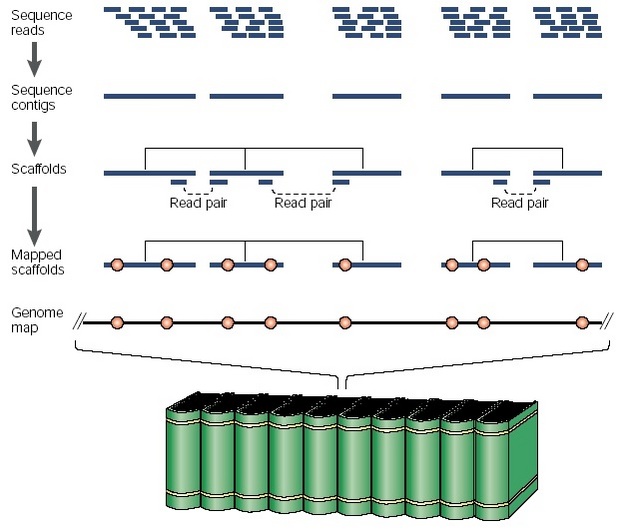
Long-range sequence assembly in whole-genome shotgun sequencing
Individual sequence reads generated in a whole-genome shotgun-sequencing project are initially assembled into sequence contigs. Groups of sequence contigs are then organized into scaffolds on the basis of linking information provided by read pairs (in each case, with one sequence read from a pair assembling into one contig and the other read into another contig). In turn, the scaffolds can be aligned relative to the source genome (represented by an encyclopedia set) by the identification of already mapped, sequence-based landmarks (e.g., STSs, genetic markers, genes; depicted as red circles) in the sequence contigs, thereby associating them with a known location on the genome map.
This image is linked to the following Scitable pages:
Scientists now have the ability to sequence complex genomes from multicellular organisms. Does the genome size correlate with the complexity of the organism? The answer is surprising.















Comments
CloseComments
Please Post Your Comment