Abstract
miR-21, which is a putative tumor onco-miR and frequently overexpressed microRNA in various tumors, has been linked to tumor progression through targeting of tumor-suppressor genes. In this study, we sought to determine whether miR-21 has any role on tumor progression of salivary adenoid cystic carcinoma (SACC) and the possible mechanisms. We found that the level of miR-21 expression was significantly higher in SACC than that in normal salivary tissues, and it is also higher in tumors with metastasis than that without metastasis. Using an anti-miR-21 inhibitor in an in vitro model, downregulation of miR-21 significantly decreased the capacity of invasion and migration of SACC cells, whereas a pre-miR-21 increased the capacity of invasion and migration of SACC cells. To explore the potential mechanisms by which miR-21 regulate invasion and migration, we identified one direct miR-21 target gene, programmed cell death 4 (PDCD4), which has been implicated in invasion and metastasis. The suppression of miR-21 in metastatic SACC-LM cells significantly increased the report activity of PDCD4 promoter and the expression of PDCD4 protein. This subsequently resulted in downregulation of the p-STAT3 protein. The level of miR-21 expression positively related to the expression of PDCD4 protein and negatively related to the expression of p-STAT3 protein in SACC specimens, respectively, indicating the potential role of the STAT3-miR-21-PDCD4 pathway in these tumors. Dysregulation of miR-21 has an important role in tumor growth and invasion by targeting PDCD4. Therefore, suppression of miR-21 may provide a potential approach for the treatment of advanced SACC patients.
Similar content being viewed by others
Main
Salivary adenoid cystic carcinoma (SACC) is one of the most common malignancy of the salivary gland, accounting for ~10% of salivary gland tumors and 30% of human salivary gland malignancies.1, 2 SACC is characterized by slow but aggressive growth, nerve and blood vessel invasion, multiple recurrences, and distant metastases.3, 4, 5 Lung metastasis and nerve metastasis are the biological characteristics of SACC.6, 7, 8 It has been reported that the incidence of SACC with distant metastasis ranged from 35 to 50% of all cases. Lung was the most common organ of its distant metastasis, and lung metastasis is still the leading death cause of patients with SACC.4, 5 Although the 5-year survival rate is high for patients with SACC, probably because of the slow growth of the tumor, the 10- and 15-year prognoses are poor because of the frequent local recurrences and distant metastases.6, 7, 8 Although multiple genetic and epigenetic alterations have been detected in SACC,3, 9, 10, 11, 12, 13 the precise molecular mechanisms of progression and metastasis of SACC remain unknown.
MicroRNAs (miRNAs) are small non-coding RNA molecules, with a length of 20–22 nucleotides, that regulate gene expression by either translational inhibition or mRNA degradation. miRNAs function as either oncogenes or tumor suppressors by inhibiting the expression of target genes, some of which are either directly or indirectly involved with canonical signaling pathways.13, 14 The roles and function of some selected miRNAs in SACC have been reported.15 Liu et al found that miR-155 facilitates cell cycle progression and promotes invasion in ACC and that the EGFR/NF-κB pathway might participate in mediating the effects of miR-155.16 Mitani et al reported that the overexpression of the miR-17 and miR-20a were significantly associated with poor outcome in SACC.17 He et al first indicate that miR-181a has an important role in the metastasis of SACC by regulating the MAPK-Snai2 pathway, and may serve as a novel therapeutic target for SACC.18 Chen et al employed microarray technology to identify miRNA expression profiles that are involved in the metastatic progression of SACC. The microarray data revealed that the levels of 38 miRNAs significantly differed between the ACC-M cells and the control ACC-2 cells. Six miRNAs (miR-4487, -4430, -486-3p, -5191, -3131, and -211-3p) were selected to validate the microarray data via qPCR. The expression of two miRNAs (miR-4487 and -4430) was significantly upregulated in the ACC-M cells, whereas the expression of two other miRNAs (miR-5191 and -3131) was significantly downregulated in the ACC-M cells.19 Sun et al reported that miR-320a inhibits metastasis in SACCs by targeting ITGB3 and may serve as a therapeutic target and prognostic marker in salivary cancers.20 However, until now, nothing has been reported about the relationship between dysregulation of miR-21 and SACC progression. The upregulation of miR-21 has been found in a variety of malignant tumors, which thus could be functioning as an oncogene in tumor developments.21 Recently, Bera et al identify a previously unrecognized signaling node where high miR-21 levels reduce rictor–programmed cell death 4 (PDCD4) interaction to increase phosphorylation of Akt and contribute to metastatic fitness of renal cancer cells.22 The relationship between miRNAs and signaling pathways in SACC is extremely complicated.
As tumor-suppressor genes, PDCD4 is also implicated in cell migration and invasion,23 suggesting that miR-21 may also have a role in invasion and metastasis. However, little is known as to how miR-21 affects these processes. The role of STAT3 in the expression of the onco-mir miR-21 is particularly important because constitutive activation of STAT3 has been demonstrated in a large number of diverse human tumors, and considerable evidence suggests that constitutive STAT3 activation actively participates in tumor formation and progression.24, 25 Abnormal STAT3 protein activation has been identified in many cancers of the breast,26 prostate,27 and pancreas,28 as well as cancers of blood-forming cells (leukemia and lymphoma).29, 30 Normally, the STAT3 protein is switched on and off in response to signals that control cell growth and development. A continuously active version of this protein relays messages to the nucleus even in the absence of these chemical signals. Researchers believe that the overactive STAT3 protein instructs cells to continue growing and dividing, and prevents damaged cells from self-destructing (undergoing apoptosis). Excess STAT3 protein may contribute to the growth of cancers by allowing abnormal cells to grow and divide uncontrollably.31, 32, 33 The present study provides the first evidence that miR-21 regulates invasion and metastasis of SACC cells, at least in part, by targeting PDCD4. Furthermore, examination of human SACC specimens revealed an inverse correlation of miR-21 with PDCD4, PDCD4 and p-STAT3.
Materials and methods
Ethics Statement
De-identified human tissue samples were obtained from the Zhejiang Province Human Tissue Specimen Bank. The use of specimens was approved by the Institutional Review Board at Zhejiang Province Cancer Hospital. Written informed consent was obtained from each patient in accordance with the requirements of our institution’s board of ethics. Twenty cases of normal salivary gland tissues (NSGs) from non-cancer patients provided written informed consent. The Institutional Review Board on Medical Ethics of Zhejiang Province Cancer Hospital also approved the method of NSG specimen collection including written informed consent from all patients.
Cell Culture and Tumor Specimens
Cell lines originated from Adenoid cystic carcinoma cells with high metastatic potential (SACC-LM) and low metastatic potential (SACC-83) were provided by Dr Sheng-Lin Li, the Peking University School of Stomatology.34 Both cell lines were cultured in RPMI 1640 complete medium with 10% inactivated FBS, 200 000 U/l penicillin, and 200 000 U/l streptomycin in humidified 5% CO2-air at 37 °C.
Thirty-seven cases of resected SACC were collected from the Department of Head and Neck Tumor Surgery, Zhejiang Province Cancer Hospital, between May 2009 and September 2010. All 37 samples were immediately placed in liquid nitrogen after removal, and 18 of 37 cases with freezing preservation had paired NSGs. All the resected specimens were also paraffin embedded and histologically examined by hematoxylin and eosin staining. The samples were collected from 16 males and 21 females, aged 35–74 years (median age 57 years). There were 19 cases of cribriform pattern carcinoma, 8 cases of salivary duct carcinoma, and 10 cases of solid carcinoma. According to the general rules of SACC staging suggested by the International Union Against Cancer,35 there were 18 cases of stage T1-3 and 19 cases of stage T4. Also, 20 cases of NSGs served as controls, in which 7 males and 13 females.
RNA Extraction
All experimental containers were treated with RNase inactivator, and all reagents were prepared in 0.11% diethypyrocarbonate-water. The tumor and their matched NSGs of 37 cases were homogenized by using a TissueLyser II (Qiagen) for 2 min at 18 Hz according to the manufacturer’s instructions at the Zhejiang Cancer Research Institute. Total RNA was isolated from the tumor and their matched NSGs using MiRNeasy Mini Kits (Qiagen, USA) and a modified acidic guanidinium phenol/chloroform method, following the manufacturer’s instructions. Total RNA was treated with DNase I (TaKaRa Bio, Otsu, Japan) to remove genomic DNA. The mRNA was purified using a poly(A) purification kit (Oligotex, Qiagen, USA) according to the manufacturer’s instructions. The quality of mRNA was assessed by the A260/280 ratio, RNA integrity was checked by denaturing agarose gel electrophoresis, and the contamination of genomic DNA was checked using PCR.
Real Time RT-PCR Analysis
PCR-based detection of mature miR-21 was performed by the TaqMan miRNA assays (ABI, Forest City, CA, USA) as described previously.36, 37 The real-time PCR results, recorded as threshold cycle numbers (Ct), were normalized against an internal control (U6 RNA), and then expressed as fold changes.36
For analysis of PDCD4 and STAT3 mRNA expression, real-time RT-PCR was performed using SYBR Premix Ex Taq™ (TaKaRa). GAPDH was used to normalize the expression levels of PDCD4 mRNA. The primer sequences for PDCD4 mRNA were as follows: (F) 5′-GGGAGTGACGCCCTTAGAAG-3′; (R) 5′-ACCTTTCTTTGGTAGTCCCCTT-3′. The primers for STAT3: (F)5′-ACCAGCAGTATAGCCGCTTC-3′; (R) 5′-GCCACAATCCGGGCAATCT-3′. The primers for GAPDH: (F) 5′-GAAGGTGAAGGTCGGAGTC-3′; (R) 5′-GAAGATGGTGATGGGATTTC-3′. All real-time RT-PCR were performed in triplicates.
Transfection
SACC-LM cells (1 × 105) were plated to 50% confluent and were transfected with 50 nmol/l GMR-miR™ miR-21 inhibitor (GenePharma, Shanghai, China) or inhibitor-negative control by Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) in Opti-MEM (Invitrogen), according to the manufacturer’s protocol. The culture medium was changed and transfection efficiency was determined using fluorescent images after transfection for 24 h. Cells were harvested for analysis after transfection for 48 h. All experiments were performed in triplicates.
Luciferase Reporter Assays
The 3′-untranslated region (UTR) of human PDCD4 mRNA containing the miR-21-binding sites was amplified by PCR from human genomic DNA and inserted into the Xba1 site of pGL3 vector (Promega, Madison, WI, USA) and named as pGL3-PDCD4-wt. Mutations in the predicted miR-21-binding sites were performed using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA), with pGL3-PDCD4-wt as a template and named as pGL3-PDCD4-mut. SACC-LM cells were co-transfected in 24-well plates with wild-type (wt) or mutant reporter plasmid vector by Lipofectamine 2000, and at 6 h after transfection, the cells were transfected again with miR-21 inhibitor or negative control. Each transfection was carried out in four wells. Luciferase assays were performed 24 h after transfection using the Dual-Luciferase reporter assay system (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity.
Transwell Migration Assay
SACC-LM cells were grown in RPMI 1640 containing 10% fetal bovine serum to ~60% confluence and 3.0 × 105 cells were transfected with miR-21 inhibitor or negative control. After 24 h, the cells were harvested by trypsinization and washed once with Hanks’ balanced salt solution (Invitrogen). To measure cell migration, 8-mm pore size culture inserts (Transwell; Costar, High Wycombe, UK) were placed into the wells of 24-well culture plates, separating the upper and the lower chambers. In the lower chamber, 400 μl of RPMI 1640 containing recombinant human hepatocyte growth factor (20 ng/ml) were added. Hepatocyte growth factor was purchased from R&D Systems (Minneapolis, MN, USA). Then, 1 μl of 105 cells were added to the upper chamber. After 24 h of incubation at 37 °C with 5% CO2, the number of cells that had migrated through the pores was quantified by counting 10 independent visual fields under the microscope (Zeiss) using a × 20 objective, and cell morphology was observed by staining with hematoxylin and eosin.
Construction of Expression Vectors
To construct a vector to express Myc-tagged PDCD4, a DNA fragment covering the PDCD4-coding region was first amplified from MCF10A cDNA by PCR (forward primer 5′-GAATTCTGGATGTAGAAAATGAGCAGA-3′ and reverse primer 5′-GCGGCCGCTCAGTAGCTCTCTGGTTTAAG-3′) and was then cloned into pCR8 (Invitrogen). After verification by DNA sequencing, the full-length PDCD4-coding fragment was finally cloned into pCMV-Myc (Clontech) at the EcoR1 and Not1 sites.
Immunohistochemical (IHC) Analysis
IHC was performed as described previously.38 Avidin–biotin–peroxidase method was used for IHC assay. All sections were deparaffinized with xylene and dehydrated with gradient alcohol followed by inactivation of endogenous peroxidase activity by 0.5% H2O2 in methanol for 10 min. Non-specific binding was blocked by incubation with 10% normal goat serum in phosphate-buffered saline for 1 h at room temperature. Then, slides were incubated with primary antibodies: anti-PDCD4 (Abcam, ab80590, Cambridge), anti-total STAT3 (Abcam ab32500, Cambridge), and anti-phospho-STAT3 (phospho Y705, Abcam, ab76315, Cambridge) antibody, respectively, at 4°C overnight, followed by biotinylated goat antimouse IgG (1:400; Sigma, St Louis, MO, USA) or goat anti-rabbit IgG (1:400; Sigma) for 1 h at room temperature. Then streptavidin–biotin–peroxidase complex assay was performed. A brown color was indicative of protein-positive expression. Peroxidase activity was developed by incubating with 0.1% 3,3-diaminobenzidine (Sigma) in phosphate-buffered saline with 0.05% H2O2 for 5 min at room temperature. The IHC staining score is determined by three independent pathologists based on combining staining frequency and intensity as previous described.38
Western Blot Analysis
Tumor specimens and their matched NSGs of 4 cases were randomly selected from 37 SACC patients for western blot analysis. Total protein was extracted and then quantified using the Lowry method.39 Western blot analysis was performed using anti-PDCD4, anti-total STAT3, and anti-phospho-STAT3 monoclonal antibodies according to the previous report9 and normalized to GAPDH (Abcam, Cambridge) protein expression.
Statistical Analysis
SPSS 17.0 statistical software (SPSS, Chicago, IL, USA) was adopted for data analysis. Counting data comparisons between groups were subjected to the χ2 test or Fisher’s exact test. miR-21 expression was shown as X±S. P<0.05 was considered statistically significant. The correlation of miR-21 and PDCD4 or p-STAT3 was analyzed by Spearman Rank Correlation, with double-sided P<0.05 as statistically significant. Survival analysis was computed by means of the Kaplan–Meier method and significance were assessed by means of the log-rank test. A univariate analysis of the Cox regression model was used to determine prognostic factors.
Results
miR-21 Is Aberrantly Overexpressed in SACC Tissue Samples and Cell Lines
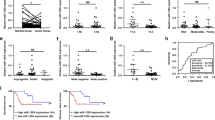
TaqMan quantitative real-time PCR analysis of miR-21 expression was succeeded in 35 (94.6%) of 37 samples. miR-21 is expressed at a significantly higher level (P=0.003; 4.8-fold on average) in SACC tissue samples than that in normal tissue samples (Figure 1a). A significant overexpression of miR-21 is also observed in two SACC cell lines (SACC-83 and SACC-LM) relative to that in normal tissue samples (SACC-83 vs NSGs, SACC-LM vs NSGs, both P=0.002). And the significant overexpression of miR-21 is found in high-metastasis-potential SACC-LM cells than that in low-metastasis-potential SACC-83 cells (Figure 1a; P=0.001). These results indicate that miR-21 expression is frequently upregulated in SACC cells, and miR-21 may have a pivotal role in SACC metastasis.
Overexpression of miR-21 in SACC tissues (SACCs) and SACC cell lines (SACC-83 and SACC-LM) as compared with normal salivary gland tissues (NSGs). (a) Comparison of expression level of miR-21 between SACCs, SACC-83, SACC-LM, and NSGs, respectively. (b) Comparison of expression level of miR-21 between SACC with metastasis and those without metastasis (non-metastasis). Relative expression level of miR-21 was determined by TaqMan RT-PCR, and all data were normalized to U6 RNA. Means±S are shown. SACC, salivary adenoid cystic carcinoma.
miR-21 Overexpression Correlates with the Metastasis of SACC Patients
The association of miR-21 expression in tumor tissues with the clinical pathologic factors of the 35 SACC patients were analyzed and are displayed in Table 1. Significant higher level of miR-21 expression was observed in the SACC of the 18 patients with metastasis than those in patients without metastasis (Figure 1b, P=0.024). No significant differences were found in miR-21 expression with respect to gender, age at surgery, tumor site, histotypes, perineural invasion, bone invasion, lymph node involvement, T phage, and distant metastasis (Table 1, all P>0.05).
miR-21 Expression Is an Independent Predictor of Survival in SACC Patients
Kaplan–Meier analysis was used to analysis the association between miR-21 expression and patients survival. As shown in Figure 2, miR-21 expression in tumor tissues was an unfavorable predictor for the survival of SACC patients after surgery. Patients with miR-21 overexpression (≥2-folds) had significantly lower disease-free survival as compared those with lower miR-21 expression (<2-folds; Figure 2 and Table 2). There was significant difference in the median survival (28 vs 64 months) between patients with miR-21 overexpression (≥2-folds) and those patients with lower miR-21 expression (<2-folds; P=0.014). Retrospective analysis using Cox regression models indicated that miR-21 overexpression in tumor tissues is a promising independent predictor of survival in SACC patients (P=0.041; Table 3). The risk ratio after adjustment for competing risk factors, sex, age, and stage of disease was found to be 7.239 (95% confidence intervals, 1.081–48.460). The results showed that increases in miR-21 expression had a greater impact on the prognosis than tumor clinical stage (Table 3).
Overexpression of miR-21 in SACC tissues correlated with poor prognosis. Cumulative disease-free survival (Cum DFS) curves are plotted against miR-21 level in SACC tissues. There was significant difference in the median survival (28 vs 64 months) between patients with miR-21 overexpression (≥2-folds) and those patients without miR-21 expression (<2-folds; P=0.014). SACC, salivary adenoid cystic carcinoma.
miR-21 Regulates SACC Cell Invasion and Migration In Vitro
In an in vitro cell invasion assay, cell invasion was significantly enhanced by transfection of MSCVpuro-miR-21 (pre-miR-21 group, 174±22 cells per high-power field (HPF); negative control group, 116±18 cells per HPF; Figure 3d–f). In contrast, cell invasion was significantly suppressed (P<0.05; ~4-fold) by transfection of an anti-miR-21 inhibitor (anti-miR-21 group, 34±12 cells per HPF; negative control group, 141±21 cells per HPF; Figure 3a–c). Similar to cell invasion assay, in cell scratch wound-healing test, overexpressed miR-21 in SACC-LM cells resulted in significant more number of the migrated cells (72±11 cells per HPF) than negative control cells (43±7 cells per HPF; Figure 4d–f; P<0.05). However, the number of SACC-LM cells migrated to the scratched area was significantly fewer (P<0.05; ~2.5-fold) in cells transfected with anti-miR-21 inhibitor (23±5 cells per HPF) than in those transfected with negative control (58±8 cells per HPF; Figure 4a–c). These results indicate that miR-21 enhances cellular capacity of invasion and migration that are associated with tumor metastasis.
miR-21 regulates cell invasion ability in SACC cells. SACC-LM cells invaded through Matrigel-coated membrane after transfection with negative control miRNA (a) and transfection with an anti-miR-21 inhibitor (b). (c) Quantification of SACC-LM cells that invaded through Matrigel-coated membrane after transfection with anti-miR-21 inhibitor or negative control. (d) SACC-83 cells that invaded through Matrigel-coated membrane after transfection with negative control vector and (e) transfection with MSCVpuro-miR-21 (pre-miR-21). (f) Quantification of SACC-83 cells that invaded through Matrigel-coated membrane after transfection with MSCVpuro-miR-21 or negative control. SACC, salivary adenoid cystic carcinoma.
miR-21 affects SACC cell migration ability. Scratch assay was used to evaluate the cell capacity of migration. (a) Migration of SACC-LM cells transfected with negative control miRNA and (b) transfected with anti-miR-21 inhibitor. (c) Quantification of SACC-LM cells that migrated from the edge of scratch in cells transfected with anti-miR-21 inhibitor or negative control. (d) Migration of SACC-83 cells transfected with negative control vector and (e) transfected with MSCVpuro- miR-21 (pre-miR-21). (f) Quantification of SACC-83 cells that migrated from the edge of scratch after transfection with MSCVpuro-miR-21 or negative control. SACC, salivary adenoid cystic carcinoma.
PDCD4 Is a Target of miR-21
To identify the potential targets of miR-21, we searched TargetScan,40 Miranda,41 PicTar,42 and MAMI (http://mami.med.harvard.edu/), for theoretical target genes whose downregulation could mediate the observed effects of miR-21. We found that PDCD4, a tumor-suppressor gene, was predicted by all the four programs as a target of miR-21. To validate whether PDCD4 is a direct target of miR-21, we constructed luciferase-reporter plasmids that contain the wild-type (wt) or mutant 3’-UTR segments of PDCD4 (Figure 5a). In addition, we mutated the miR-21-binding site in the 3’UTR of PDCD4 (in the reporter plasmid). The wt or mutant reporter plasmid was co-transfected into SACC-LM cells along with miR-21 inhibitor or negative control. Compared with negative control, miR-21 inhibitor significantly increased the relative luciferase activity when co-transfected with the wt reporter plasmid. However, miR-21 inhibitor-mediated increase of luciferase activity was abolished in the mutant reporter plasmid (Figure 5b). These results indicate that PDCD4 is a direct target of miR-21 and that miR-21 suppresses PDCD4 by direct binding to the 3’-UTR of PDCD4.
The 3′-UTR of PDCD4 mRNA is a target for miR-21. (a) Predicted miR-21-binding sites within the 3′-UTR of PDCD4 mRNA. The PDCD4 UTR sequence reveals a putative mir-21-binding site (http://www.mirbase.org/index.shtml). The underlined sequence denotes a deletion in Luc-PDCD4-UTR-d. (b) The wild-type (wt) or mutant (mut) reporter plasmid was co-transfected into SACC-LM cells with miR-21 inhibitor or negative control (NC). The normalized luciferase activity in the control group was set as relative luciferase activity 1. Luciferase activity of pGL3-PDCD4-wt was significantly increased by miR-21 inhibitor (*P<0.05). However, luciferase activity of pGL3-PDCD4-mut was not affected by miR-21 inhibitor (P>0.05). 3’-UTR, 3’-untranslated region.
Then we assessed whether the down-expression of miR-21 affected PDCD4 protein expression in SACC cells. SACC-LM cells were transiently transfected with miR-21 inhibitor. The expression of PDCD4 protein was significantly increased when endogenous miR-21 was knocked down by the miR-21 inhibitor. Because Stat3 has been reported as a downstream target of PDCD4, therefore, we checked the protein expression of the phosphorylated STAT3 expression and found that the expression of the phosphorylated STAT3 was concomitantly decreased by miR-21 inhibitor (Figure 6a). However, PDCD4 and STAT3 mRNA were almost not affected by the alternation of miR-21 levels (Figure 6c). Interestingly, the expression of PDCD4 protein was significantly increased, and the expression of miR-21 levels was concomitantly decreased when the knockdown of endogenous phosphorylated STAT3 expression by RNA interference, indicating that there may be a feedback loop among miR-21-PDCD4-p-Stat3 (Figure 6d2,e and f).
miR-21 regulates PDCD4 expression at the post-transcriptional level and influences the phosphorylation of STAT3. (a) Cells were transfected with miR-21 inhibitor, inhibitor-negative control (NC), or blank control culture medium (MOCK). Cell lysates were obtained after 48 h for analysis. miR-21 expression was detected by TaqMan RT-PCR. The results were normalized to U6 expression and expressed as fold change relative to the corresponding NC (*P<0.05). (b, d) The expression of PDCD4 and phosphorylated STAT3 were examined by western blot analysis. The results were normalized to GAPDH protein expression and expressed as fold change relative to the corresponding NC (*P<0.05). (c) The expression of PDCD4 and STAT3 mRNA were examined by real-time RT-PCR. The results were normalized to GAPDH mRNA expression and expressed as fold change relative to the corresponding NC. (e) The silencing of p-STAT3 regulates miR-21 expression. Cells were transfected with shRNA-p-STAT3, shRNA-NC, or blank control culture medium. Cells were obtained after 48 h for analysis. miR-21 expression was detected by TaqMan RT-PCR. The results were normalized to U6 expression and expressed as fold change relative to the corresponding NC (*P<0.05). (f) The expression of PDCD4 and phosphorylated STAT3 was examined by western blot analysis after cells treated with shRNA-p-STAT3. The results were normalized to GAPDH protein expression and expressed as fold change relative to the corresponding NC (*P<0.05). d1: PDCD4 protein was significantly increased and phosphorylated STAT3 protein was concomitantly decreased when endogenous miR-21 was knocked down by the miR-21 inhibitor; d2: PDCD4 protein was significantly increased and phosphorylated STAT3 protein was concomitantly decreased when interfered by shRNA-p-STAT3. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
PDCD4 Protein Level Negatively Correlates with miR-21 and p-STAT3 Protein Expression in Tumor Specimens
The PDCD4 and p-STAT3 protein in all 35 paraffin-embedded samples were analyzed by IHC staining. In comparison with their matched NSGs, tumor tissues expressed significantly lower levels of PDCD4 protein (P<0.0001, χ2-test), and significantly higher levels of p-STAT3 protein (P<0.0001, χ2-test). The expression of PDCD4 and p-STAT3 protein was validated by western blotting, and representative examples were shown in Figure 6a. The miR-21 levels negatively correlated to PDCD4 protein and positively correlated to p-STAT3 protein (Figure 7a and b). To determine the exact association, we used the Pearson’s method for statistical analysis. We confirmed that the inverse correlation between miR-21 levels and PDCD4 protein (r=–0.556, P=0.001, Figure 7c), positive correlation between miR-21 levels and p-STAT3 protein (r=0.661, P=0.000, Figure 7d), in these SACC tissues. We also confirmed the inverse correlation between PDCD4 protein and p-STAT3 protein in 35 SACC specimens (r=−0.972, P=0.000, Figure 7e).
Expression of PDCD4, p-STAT3, and miR-21 in matched SACC tumor specimens. (a) Expression of PDCD4, STAT3, and p-STAT3 protein in four pairs of matched SACC tissues and normal tissues, as detected by western blotting and normalized to β-actin protein expression. The specimen number is shown above each pair comprising tumor (T) and normal (N). The miR-21 expression folds of the four cases were shown under the western blotting panels. (b) Relative expression of PDCD4 and p-STAT3 protein in SACC tissues were detected by immunohistochemistry. Original magnification × 200. Case 1: Absent of PDCD4 protein expression and high expression of p-STAT3 protein in a SACC tissue with miR-21 overexpression. Case 4: a representative positive, positive expression of PDCD4 and p-STAT3 protein in a SACC tissue without miR-21 overexpression. (c) Correlation between miR-21 expression and PDCD4 protein levels in SACC tissues (Pearson correlation, r=−0.556, P=0.001). (d) Correlation between miR-21 expression and p-STAT3 protein levels in SACC tissues (Pearson correlation, r=0.661, P=0.000). (e) Correlation between PDCD4 protein and p-STAT3 protein levels in SACC tissues (Pearson correlation, r=−0.972, P=0.000). GAPDH, glyceraldehyde-3-phosphate dehydrogenase; SACC, salivary adenoid cystic carcinoma.
In a word, we found that high levels of miR-21 in SACC samples correlate with low level of PDCD4 protein and high level of p-STAT3 protein.
Discussion
Metastasis is a major cause of cancer-related death. Previous research elucidated that miRNAs have an important role in the occurrence, development, and metastasis of SACC.16, 17, 18, 19, 20 Although miR-21 do not be included these upregulated miRNAs identified by Chen et al,19 it is possible that bounded by selected cell line. The identification of miRNAs in highly metastatic SACC cells may provide an improved understanding of the mechanisms involved in metastatic progression, which would aid in the development of novel strategies for the treatment of SACC. Hence, identifying the role of miR-21 in invasion and metastasis has direct clinical implications. Although it is now known that miR-21 might has a key role in cancer development, the underlying mechanism is largely unknown. We show here that miR-21 is significantly upregulated in SACC and the dysregulation of it may be associated with tumor invasion, migration, and metastasis. And this effect of miR-21 in SACC probably through downregulation of its direct target of PDCD4 and this results in subsequent activation of p-Stat3.
In this study, we found that miR-21 expression was significantly higher in metastatic SACC than that in non-metastatic SACC samples. We also found that miR-21 remarkably increased invasion and migration in SACC cells, these results are consistent with previous findings from other cancers.21, 22, 43, 44 In addition, we showed that knockdown of miR-21 could dramatically decrease cell invasion and migration in SACC cells. Overexpression of miR-21 has been reported in various human cancers. All these results including our data indicate that miR-21 has multiple functions in diverse cancers and could be recognized as an important onco-miR.14, 21 Therefore, targeting miR-21 may be a potential therapeutic strategy for these tumors.
It has been suggested that PDCD4 is a potential target for miR-21,45 and PDCD4 expression might have an essential role in the progression of SACC.46 We confirmed that PDCD4 is a direct target for miR-21 through the miR-21-binding site at the 3′-UTR. PDCD4 has been reported as a tumor-suppressor gene in various tumors.22, 47, 48 It performs an important function in tumor invasion and metastasis through inhibition of STAT3 activation, which are known to be involved in cancer progression.22, 47, 48, 49 Our data suggested that PDCD4 expression has a negative correlation with the expression of miR-21 in SACC tissues and cell lines at protein level. Applying specific inhibitor of miR-21 can rescue PDCD4 expression, negatively regulated SACC cell invasion and metastasis. The results suggest that upregulation of miR-21 was attributive to, at least in part, the downregulation of metastasis-related tumor suppressors PDCD4, although more samples may be required to make a firm conclusion. Because constitutive activation of STAT3 has been demonstrated in a large number of diverse human tumors, and considerable evidence suggests that constitutive STAT3 activation actively participates in tumor formation and progression,24, 25 we also studied the expression of p-stat3 in our in vitro model and in SACC samples. We found that p-Stat3 expression is significantly upregulated in SACC samples and its expression was inversely correlated with PDCD4 expression but positively correlated with miR-21 level. In addition, silencing of p-STAT3 downregulated the expression of miR-21 and upregulated the expression of PDCD4 protein and resulted in the inhibition of invasion and metastasis in SACC-LM cells. We verified in vitro that miR-21 negatively regulates PDCD4 and positively regulates p-STAT3, which in turn alters miR-21 expression, suggesting that the STAT3-miR-21-PDCD4 feedback regulatory network may have an important role in SACC pathogenesis and progression. Therefore, the decrease of miR-21 with miR-21 inhibitor in SACC-LM cells could target PDCD4 gene and in turn enhanced PDCD4 protein expression, then suppressed p-STAT3 protein expression, further feedback reduced miR-21 expression, and the final leading to the inhibition of cell invasion and migration.
miR-21 is overexpressed in SACC cells, and an aberrant expression of miR-21 can alter multiple biological processes of human SACC cells, such as proliferation, invasion, and migration, probably through regulating PDCD4 and other critical target genes. miR-21 can serve as a biomarker for patients with SACC, and an inhibitory strategy against the STAT3-miR-21-PDCD4 pathway will have a strong rationale for the treatment of SACC patients.
References
Moskaluk A . CA. Adenoid cystic carcinoma: clinical and molecular features. Head Neck Pathol 2013;7:17–22.
Liu J, Shao C, Tan ML et al. Molecular biology of adenoid cystic carcinoma. Head Neck 2012;34:1665–1677.
He JF, Ge MH, Zhu X et al. Expression of RUNX3 in salivary adenoid cystic carcinoma: implications for tumor progression and prognosis. Cancer Sci 2008;99:1334–1340.
Bradley PJ . Adenoid cystic carcinoma of the head and neck: a review. Curr Opin Otolaryngol Head Neck Surg 2004;12:127–132.
Hu K, Gan YH, Li SL et al. Relationship of activated extracellular signal-regulated kinase 1/2 with lung metastasis in salivary adenoid cystic carcinoma. Oncol Rep 2009;21:137–143.
Zhang CY, Mao L, Li L et al. Promoter methylation as a common mechanism for inactivating E-cadherin in human salivary gland adenoid cystic carcinoma. Cancer 2007;110:87–95.
Shi H, Wang J, Dong F et al. The effect of proteoglycans inhibited by RNA interference on metastatic characters of human salivary adenoid cystic carcinoma. BMC Cancer 2009;9:456.
Lagha A, Chraiet N, Ayadi M et al. Systemic therapy in the management of metastatic or advanced salivary gland cancers. Oral Oncol 2012;48:948–957.
Ge MH, Chen C, Xu JJ et al. Critical regions and spreading of runt-related transcription factor-3 C-phosphate-G (CpG) island methylation in human salivary gland adenoid cystic carcinoma. Hum Pathol 2011;42:1862–1872.
Ge MH, Chen C, Xu JJ et al. Unfavorable clinical implications for hypermethylation of RUNX3 in patients with salivary gland adenoid cystic carcinoma. Oncol Rep 2011;26:349–357.
Rutherford S, Hampton GM, Frierson HF et al. Mapping of candidate tumor suppressor genes on chromosome 12 in adenoid cystic carcinoma. Lab Invest 2005;85:1076–1085.
Stallmach I, Zenklusen P, Komminoth P et al. Loss of heterozygosity at chromosome 6q23-25 correlates with clinical and histologic parameters in salivary gland adenoid cystic carcinoma. Virchows Arch 2002;440:77–84.
Bernheim A, Toujani S, Saulnier P et al. High-resolution array comparative genomic hybridization analysis of human bronchial and salivary adenoid cystic carcinoma. Lab Invest 2008;88:464–473.
Davis BN, Hata A . microRNA in Cancer—The involvement of aberrant microRNA biogenesis regulatory pathways. Genes Cancer 2010;1:1100–1104.
Nicolas FE, Lopez-Martinez AF . MicroRNAs in human diseases. Recent Pat DNA Gene Seq 2010;4:142–154.
Liu L, Hu Y, Fu J et al. MicroRNA155 in the growth and invasion of salivary adenoid cystic carcinoma. J Oral Pathol Med 2013;42:140–147.
Mitani Y, Roberts DB, Fatani H et al. MicroRNA profiling of salivary adenoid cystic carcinoma: association of miR-17-92 upregulation with poor outcome. PLoS One 2013;8:e66778.
He Q, Zhou X, Li S et al. MicroRNA-181a suppresses salivary adenoid cystic carcinoma metastasis by targeting MAPK-Snai2 pathway. Biochim Biophys Acta 2013;1830:5258–5266.
Chen W, Zhao X, Dong Z et al. Identification of microRNA profiles in salivary adenoid cystic carcinoma cells during metastatic progression. Oncol Lett 2014;7:2029–2034.
Sun L, Liu B, Lin Z et al. MiR-320a acts as a prognostic factor and Inhibits metastasis of salivary adenoid cystic carcinoma by targeting ITGB3. Mol Cancer 2015;14:96.
Selcuklu SD, Donoghue MT, Spillane C . miR-21 as a key regulator of oncogenic processes. Biochem Soc Trans 2009;37:918–925.
Bera A, Das F, Ghosh-Choudhury N et al. microRNA-21-induced dissociation of PDCD4 from rictor Contributes to Akt-IKKβ-mTORC1 axis to regulate select renal Cancer cell invasion. Exp Cell Res 2014;328:99–117.
Zhu S, Wu H, Wu F et al. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res 2008;18:350–359.
Turkson J, Jove R . STAT proteins: novel molecular targets for cancer drug discovery. Oncogene 2000;19:6613–6626.
Bowman T, Garcia R, Turkson J et al. STATs in oncogenesis. Oncogene 2000;19:2474–2488.
Wei W, Tweardy DJ, Zhang M et al. STAT3 signaling is activated preferentially in tumor-initiating cells in claudin-low models of human breast cancer. Stem Cells 2014;32:2571–2582.
Bishop JL, Thaper D, Zoubeidi A . The multifaceted roles of STAT3 signaling in the progression of prostate cancer. Cancers (Basel) 2014;6:829–859.
Panni RZ, Sanford DE, Belt BA et al. Tumor-induced STAT3 activation in monocytic myeloid-derived suppressor cells enhances stemness and mesenchymal properties in human pancreatic cancer. Cancer Immunol Immunother 2014;63:513–528.
Ge Y, Yang B, Xu X et al. Cryptotanshinone acts synergistically with imatinib to induce apoptosis of human chronic myeloid leukemia cells. Leuk Lymphoma 2014;25:1–9.
Munoz J, Dhillon N, Janku F et al. STAT3 inhibitors: finding a home in lymphoma and leukemia. Oncologist 2014;19:536–544.
Qi QR, Yang ZM . Regulation and function of signal transducer and activator of transcription 3. World J Biol Chem 2014;5:231–239.
Xiong A, Yang Z, Shen Y et al. Transcription factor STAT3 as a novel molecular target for cancer prevention. Cancers (Basel) 2014;6:926–927.
Zhang HF, Lai R . STAT3 in cancer-friend or foe? Cancers (Basel) 2014;6:1408–1440.
Hu K, Li SL, Gan YH et al. Epiregulin promotes migration and invasion of salivary adenoid cystic carcinoma cell line SACC-83 through activation of ERK and Akt. Oral Oncol 2009;45:156–163.
International Union Against Cancer (UICC). In: Sobin LH, Gospodarowicz MK, Ch Wittekind (eds). TNM Classification of Malignant Tumors, 7th edn. Wiley-Liss: New York, 2010.
Chen C, Ridzon DA, Broomer AJ et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 2005;33:e179.
Lao K, Xu NL, Yeung V et al. Multiplexing RT-PCR for the detection of multiple miRNA species in small samples. Biochem Biophys Res Commun 2006;343:85–89.
Ling ZQ, Guo W, Lu XX et al. A Golgi-specific protein PAQR3 is closely associated with the progression, metastasis and prognosis of human gastric cancers. Ann Oncol 2014;25:1363–1372.
Ling ZQ, Lv P, Lu XX et al. Circulating methylated XAF1 DNA indicates poor prognosis for gastric cancer. PLoS One 2013;8:e67195.
Lewis BP, Burge CB, Bartel DP . Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005;120:15–20.
Krek A, Grün D, Poy MN et al. Combinatorial microRNA target predictions. Nat Genet 2005;37:495–500.
John B, Enright AJ, Aravin A et al. Human MicroRNA targets. PLoS Biol 2004;2:e363.
Li P, Mao WM, Zheng ZG et al. Down-regulation of PTEN expression modulated by dysregulated miR-21 contributes to the progression of esophageal cancer. Dig Dis Sci 2013;58:3483–3493.
Zhu W, Xu B . MicroRNA-21 identified as predictor of cancer outcome: a meta-analysis. PLoS One 2014;9:e103373.
Roldo C, Missiaglia E, Hagan JP et al. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J Clin Oncol 2006;24:4677–4684.
Qi C, Shao Y, Li N et al. Prognostic significance of PDCD4 expression in human salivary adenoid cystic carcinoma. Med Oncol 2013;30:491.
Matsuhashi S, Hamajima H, Xia J et al. Control of a tumor suppressor PDCD4: Degradation mechanisms of the protein in hepatocellular carcinoma cells. Cell Signal 2014;26:603–610.
Hwang SK, Baker AR, Young MR et al. Tumor suppressor PDCD4 inhibits NF-κB-dependent transcription in human glioblastoma cells by direct interaction with p65. Carcinogenesis 2014;35:1469–1480.
Yang CH, Yue J, Pfeffer SR et al. MicroRNA miR-21 regulates the metastatic behavior of B16 melanoma cells. J Biol Chem 2011;286:39172–39178.
Acknowledgements
This research was partly supported by Zhejiang Provincial Program for the Cultivation of High-level Innovative Health talents (Ge M-H and Ling Z-Q), Zhejiang Province Natural Science Foundation (No. LY14H160014), National Natural Science Foundation of China (No. 81202127), and a key project of Zhejiang Provincial Medicine and Hygiene Platform Programs (No. 2012ZDA005).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declared no conflict of interest.
Rights and permissions
About this article
Cite this article
Jiang, LH., Ge, MH., Hou, XX. et al. miR-21 regulates tumor progression through the miR-21-PDCD4-Stat3 pathway in human salivary adenoid cystic carcinoma. Lab Invest 95, 1398–1408 (2015). https://doi.org/10.1038/labinvest.2015.105
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.2015.105
This article is cited by
-
MiR-744-5p inducing cell death by directly targeting HNRNPC and NFIX in ovarian cancer cells
Scientific Reports (2018)
-
MiR21 sensitized B-lymphoma cells to ABT-199 via ICOS/ICOSL-mediated interaction of Treg cells with endothelial cells
Journal of Experimental & Clinical Cancer Research (2017)
-
miR-21 Might be Involved in Breast Cancer Promotion and Invasion Rather than in Initial Events of Breast Cancer Development
Molecular Diagnosis & Therapy (2016)