Abstract
The present study retrospectively examined the correlation between the outcome of patients with locally advanced oesophageal squamous cell carcinoma (cT3-4 cN0-1 cM0) after multimodal treatment (radiochemotherapy±surgical resection), and the presence of genetic polymorphisms in genes involved in folate metabolism. In total, 68 patients who took part in a prospective multicentric trial received 5-fluorouracil (FU)-based radiochemotherapy, optionally followed by surgery. DNA was extracted from pretherapeutic tumour biopsies and was subsequently genotyped for common genetic polymorphisms of three genes (MTHFR C677T, MTR A2756G, TS tandem repeat polymorphism) involved in folate metabolism and potentially in sensitivity to 5-FU-based chemotherapy. The genotypes were correlated with tumour response to polychemotherapy, radiochemotherapy and with overall survival. Tumours with the MTR wild-type genotype (2756AA) showed a median survival time of 16 months, whereas tumours with an MTR variant genotype (2756AG/2756GG) showed a median survival time of 42 months (P=0.0463). No prognostic impact could be verified for the genotypes of the MTHFR genes and the TS gene. Among tumours treated with radiochemotherapy and subsequent resection, MTR variant genotype showed higher histopathological response rate than tumours with MTR wild-type genotype (P=0.0442). In contrast, no significant relationship between clinically determined tumour regression after polychemotherapy and polymorphisms of the three genes under analysis was observed. In conclusion, pretherapeutic determination of the MTR A2756G polymorphism may predict survival of multimodally treated oesophageal squamous cell carcinomas. Determination of MTHFR C677T and TS tandem repeat polymorphism has no predictive value.
Similar content being viewed by others
Main
In recent years, the prognosis of patients with oesophageal squamous cell carcinoma has only slightly improved. The results of surgical therapy have been poor, with 5-year survival rates varying between 9 and 40%, even in resectable stages (Enzinger and Mayer, 2003). Therefore, combined treatment modalities including chemotherapy, irradiation and surgical treatment were investigated in an increasing number of studies in order to improve survival of oesophageal cancer patients. These studies have indicated a complete response (CR) in 20–40% of patients preoperatively treated with combined radiotherapy and chemotherapy (Forastiere et al, 1997). However, concerning survival, proven benefit from combined neoadjuvant treatment modalities has been unequivocally shown only for the subset of patients showing a CR at histopathologic examination (Lerut et al, 1999; Ancona et al, 2001; Brücher et al, 2004). It may, therefore, be of great interest to find parameters that may help to identify those patients who will benefit from multimodal treatment modalities prior to the start of therapy (Forastiere et al, 1997; Lerut et al, 1999).
Methylenetetrahydrofolate reductase (MTHFR) and methionine synthase (MTR) are key enzymes in folate metabolism. The substrate for MTHFR, 5,10-methylene-tetrahydrofolate (THF) is used for conversion of dUMP to dTMP by the enzyme thymidylate synthase (TS). The enzyme product of MTHFR, 5-methyl-THF, serves as carbon donor for the remethylation of homocysteine to methionine, which is catalysed by the enzyme MTR (Figure 1) (Choi et al, 1998; Balley and Gregory, 1999). The MTHFR gene is highly polymorphic in the general population. The major nucleotide 677 polymorphism (C to T) results in an alanine to valine substitution, which alters enzyme activity (Goyette et al, 1994). Thus, individuals with the variant 677TT genotype have about 30% of the in vitro MTHFR enzyme activity of those with the 677CC wild-type genotype, whereas heterozygotes (677CT) have about 65% enzyme activity (Balley and Gregory, 1999). The Methionine Synthase (MTR) gene contains a common polymorphism at bp 2756 (MTR A2756G), which causes an amino-acid substitution from aspartic acid to glycine at codon 919 (D919G). Individuals with the 2756GG genotype have lower plasma homocysteine (or higher folate) levels than those with the 2756AA genotype (Tsai et al, 2000; Chen et al, 2001), indicating an increased enzymatic activity of the variant genotype.
An accumulation of 5,10-methylene-THF resulting from the MTHFR C677T polymorphism and/or the MTR A2756G polymorphism may have an effect on the response of cancer cells to 5-fluorouracil (5-FU), a chemotherapeutic agent frequently used in the neoadjuvant treatment of oesophageal squamous cell carcinoma. This is because 5-FU can form a ternary complex involving 5-fluoro-2′-deoyuridine-5′-monophosphate (5-FdUMP), TS, and 5,10-methylene-THF. The formation of this complex inhibits TS activity, which subsequently depletes intracellular thymidylate levels and ultimately suppresses DNA synthesis. Therefore, the MTHFR C677T polymorphism or the MTR A2756G polymorphism, which increase the intracellular concentrations of 5,10-methylene-THF, may increase the cytotoxic effect of 5-FU by increasing the formation of the 5,10-methylene-THF-TS-5-FdUMP ternary complex. The potential relevance of this mechanism has been recently shown in vitro where in colon cancer cell lines the MTHFR 677T polymorphism was associated with increased to sensitivity to 5-FU treatment (Sohn et al, 2004). Moreover, patients with advanced colorectal bearing the polymorphic MTHFR allele (either heterozygous or homozygous) responded better to 5-FU-based chemotherapy than patients with the wild-type MTHFR allele (Cohen et al, 2003). In contrast, the role of the MTHFR and MTR polymorphisms in 5-FU-treated oesophageal cancer has not been determined so far.
Additionally, the TS gene also contains a genetic polymorphism that may be also involved in efficacy of 5-FU-based chemotherapy. It is a 28-bp tandem repeat sequence within the 5′-untranslated region, and the vast majority of individuals show one of three genotypes, that is, two tandem repeats (2R/2R), three tandem repeats (3R/3R) or a heterozygous (2R/3R) genotype (Horie et al, 1995). This tandem repeat seems to function as an enhancer element since in vitro studies have shown that stepwise increase of TS mRNA expression and TS enzyme activity are associated with increasing number of tandem repeat sequences. Indeed, there is growing evidence that in colorectal cancer the TS 2R/2R genotype is associated with improved response and survival following 5-FU-based chemotherapy (Iacopetta et al, 2001; Villafranca et al, 2001; DiPaolo and Chu, 2004).
Therefore, in the present study, genotypes of MTHFR, MTR and TS were determined in preoperative biopsies derived from of multimodally treated patients with locally advanced oesophageal squamous cell carcinoma. Subsequently, these findings were correlated with response to 5-FU-based chemotherapy and overall survival.
Material and methods
Patients
All patients of the present investigation were part of a prospective multicentric phase III study and gave written, informed consent to participate in this study. The results of this study, together with the criteria for patient selection and study design, have been extensively described elsewhere (Stahl et al, 1996, 2005).
Patient selection and study design
In total, 68 patients with locally advanced oesophageal cancer (i.e. category T3/T4 according to the UICC classification (Sobin and Wittekind, 1997) defined by CT scan and endoscopic ultrasound), with or without regional lymph node metastases, were eligible.
Three courses of chemotherapy were administered within 9 weeks, followed by 4 weeks of radiotherapy with concomitant chemotherapy during the first 7 days. After the administration of chemotherapy and a cumulative dose of 40 Gy of radiotherapy, patients were either treated by at least further 25 Gy of radiotherapy (definitive radiochemotherapy) or by transthoracal oesophageal resection. Postoperative treatment was not performed.
Preoperative chemotherapy and preoperative radiochemotherapy
Chemotherapy consisted of folinic acid 300 mg m−2, etoposide 100 mg m−2, bolus 5-FU 500 mg m−2, and cisplatin 30 mg m−2 on days 1–3, every 3 weeks (FLEP protocol).
Combined radiochemotherapy was administered on days 22–28 of the last course of chemotherapy. The oesophagus was irradiated using parallel-opposed anterior and posterior fields and photons from a 10- to 15-MV linear accelerator. A total dose of 40 Gy was given in daily fractions of 2 Gy, five times per week. During the first days of irradiation, the following chemotherapy was administered: cisplatin 50 mg m−2 on days 2 and 8, and etoposide 100 mg m−2 on days 4–6.
Surgery
Resection of the oesophagus and the proximal stomach was performed by a combined right thoracal and abdominal approach. Resection included excision of the paraoesophageal, paracardial, left gastric, and celiac lymph nodes.
Criteria for response to chemotherapy and radiochemotherapy
Response to chemotherapy was evaluated clinically after the third course and included barium oesophagogram, oesophagoscopy, and computed tomography of the chest and abdomen. Response was categorised as follows: CR – normal barium oesophagogram, no visible tumour by oesophagoscopy, biopsies free of tumour tissue, and normal CT; partial response (PR) – greater than 50% tumour regression as evaluated by CT, and greater than 50% reduction of intraoesophageal tumour extension as assessed by barium swallow and oesophagoscopy; no change (NC) – less than 50% regression of tumour extension, and no evidence of tumour progression; progressive disease (PD) – increasing tumour obstruction indicated by barium swallow or oesophagoscopy, and increasing tumour diameter assessed by CT.
Response to radiochemotherapy was evaluated by histopathological examination of cases that underwent transthoracal oesophageal resection. In this evaluation, complete responders (no viable tumour cells in the resection specimens) were separated from cases were incomplete or no tumour regression was detectable.
Pathologic review of pretherapeutic tumour biopsies
Tumour biopsies that had been endoscopically obtained during pretherapeutic staging procedures were retrieved from the files of pathologic institutes associated with the medical centers that took part in this study. Of the 68 patients, 60 were male and eight were female. The median age was 57 years (range: 37–70). A total of 58 tumours had been clinically categorised as T3 and 10 tumours had been categorised as T4; 15 cases were in category N0 and 53 cases were in category N1. Histologic slides of the biopsy specimens were stained with hematoxylin and eosin for the determination of tumour type and tumour grade according to the WHO criteria. Three tumours were graded as G1, 34 tumours were graded as G2, and 31 tumours were graded as G3.
Genotyping of MTHFR C677T, MTR A2756G and TS
For the determination of individual genotypes, genomic DNA was extracted from formaldehyde-fixed tumour biopsies using commercially available DNA-preparation kits (Qiagen, Hilden, Germany). The MTHFR C677T genotype was determined by real-time fluorescence PCR and melting curve analysis using a LightCycler instrument (Roche Diagnostics, Mannheim, Germany). PCR conditions, primers and probes and melting curve analysis were applied as previously described (Sarbia et al, 2005). A melting curve with a single peak at low melting temperature (50°C) indicates a homozygous variant (677TT) genotype, a single peak at high melting temperature (60°C) indicates a homozygous wild-type (677CC) genotype, and a double peak indicates a heterozygous genotype.
A PCR-RFLP assay was used to determine the MTR A2756G genotype. The primers were forward 5′-GAA CTA GAA GAC AGA AAT TCT CTA-3′ and reverse 5′-CAT GGA AGA ATA TGA AGA TAT TAG A-3′ (Leclerc et al, 1996). The PCR product is digested with HaeIII (New England Biolabs, Beverly, MA, USA); and the amplified fragment of 189 bp is cut into fragments of 159 and 30 bp in the presence of the G allele.
The promoter region of the TS gene was amplified by PCR using the following primers: forward 5′-AAAAGGCGCGCGGAAGGGGTCCT-3′ and reverse 5′-TCCGAGCCGGCCACCAGGCAT-3′ (Iacopetta et al, 2001). The PCR products were analysed by length differences using ethidium-bromide-stained 2% agarose gels.
Statistical analysis
SAS software package (SAS Institute Inc., Carey, NC, USA) was used for statistical analyses. Statistical analysis of the correlation between tumour regression following chemotherapy and genotypes (MTHFR, MTR, TS) was performed by means of the two-tailed Fisher's exact test. Therefore, the parameters were dichotomised as follows: tumour response (CR/PR vs NC/PD), MTHFR (CC vs CT/TT), MTR (AA vs AG/GG) and TS (2R/2R vs 2R/3R/3R/3R). The Kaplan–Meier method for analysis of censored data was used for calculating survival rates and tested with the log-rank test and the Wilcoxon test. Probabilities <0.05 were regarded as statistically significant.
Results
Correlation between genotypes and tumour regression following chemotherapy and radiochemotherapy
Following FLEP chemotherapy, three patients (4.4%) received CR, 28 PR (41.2%), 32 NC (47.1%) and five PD (7.3%). Data on histopathological response evaluation after radiochemotherapy was available only in a subset of patients that underwent transthoracal oesophageal resection (n=20). Among these cases, seven (35.0%) showed complete regression without viable residual tumour cells, whereas in the other 13 (65.0%) cases either incomplete regression or no histological signs of tumour regression were present.
In total, 67 samples could be genotyped successfully for the MTHFR gene (98.5%), 64 for the MTR gene (94.1%) and 57 for the TS gene (83.8%).
No significant relationship between tumour regression after FLEP chemotherapy and polymorphisms of the three genes under analysis was observed (Table 1). In contrast, response to radiochemotherapy was correlated with the MTR gene polymorphism. Tumours with an MTR variant genotype (2756AG/2756GG) showed complete histopathological regression in 46.2% of cases whereas tumours with the MTR wild-type genotype (2756AA) showed complete regression in 0% of cases (Table 2). (P=0.0442 according to the χ2 test). No correlation between response to radiochemotherapy and MTHFR and TS genotypes was found.
Survival analysis
The overall survival of all 68 patients has been followed up every 3 months for the first 3 years after the end of treatment; afterwards every 6 months. One patient was lost to follow-up. At the end of the follow-up period (30.4.2005), 14 of the remaining 67 patients (20.9%) were still alive. The follow-up time for all 67 patients ranged from 4 to 114 months (median: 22 months). The follow-up time for the 14 patients at risk ranged from 29 to 114 months (median: 83 months).
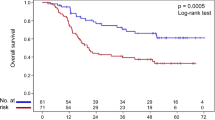
All patients received complete treatment. Of the 67 patients suitable for survival analyses, 38 received definitive radiochemotherapy and 29 were treated by radiochemotherapy and subsequent oesophageal resection. Analysing all 67 patients, tumours with the MTR wild-type genotype (2756AA) showed a median survival time of 16 months whereas tumours with an MTR variant genotype (2756AG/2756GG) showed a median survival time of 42 months (Figure 2) (P=0.0463 according to the Wilcoxon test; P=0.566 according to the log-rank test).
Survival curves of oesophageal SCC patients treated either by neodjuvant radiochemotherapy and subsequent resection or by definitive radiochemotherapy in relation Methionine Synthase genotype (n=63, A). Subgroup analysis for patients either treated by neoadjuvant radiochemotherapy and resection (n=28, B) or by definitive radiochemotherapy (n=35, C).
No prognostic impact could be verified for the genotypes of the MTHFR genes and the TS gene either in the group as a whole (Table 3) or when stratified, according to the type of therapy (radiochemotherapy followed by resection or definitive radiochemotherapy).
Discussion
The current study indicates that multimodally treated oesophageal squamous cell carcinoma patients with the MTR 2756AG/2756GG genotypes may have better survival than individuals with the MTR 2756AA genotype. Mechanistically, this may make sense taking into account the putative effect of this polymorphism on folate metabolism. Thus, MTR catalyses the transfer of the methyl base from 5-methyl THF to homocysteine, thereby producing methionine and THF. THF is the precursor of 5′-formyl THF, which is an inhibitor of thymidylate synthase in cooperation with 5′-FdUMP, which is a metabolite of 5′-FU. Therefore, an increased activity of the MTR gene may lead to an accumulation of 5′-formyl THF and thereby to a pronounced cytotoxic effect of 5′-FU treatment. The MTR 2756GG polymorphism was initially thought to be associated with lower enzyme activity than the MTR 2756AA genotype (Leclerc et al, 1996). However, more recent investigations indicate that the variant genotypes (2756AG, 2756GG) may be associated with lower homocysteine and/or higher methionine levels than the wild-type genotype, implying a more effective enzyme activity (Tsai et al, 2000; Chen et al, 2001; Silaste et al, 2001).
On the other hand, we did not observe any effect of the MTHFR C677T polymorphism on survival. If the hypothesis is true that accumulation of 5′-formyl THF increases the effect of 5′-FU-based therapy, this observation is difficult to understand. This is because the variant genotype of the MTHFR gene is characterised by less effective catalyzation of 5′10′-methylene THF to 5′methyl THF and thus to an accumulation of 5′-formyl THF. However, one clearly has to take into account that a large variety of other factors than genetic polymorphisms may influence the activity of the key enzymes involved in folate metabolism. Among others, enzyme activities may be influenced by transcriptional regulation, post-transcriptional modification and protein modification. Moreover, 5-FU was not the only cytotoxic drug used in the chemotherapy protocol of the current study. Therefore, the putative effect of 5-FU-associated polymorphisms may be superposed by other effects. Therefore, currently, only empirical studies like the present one may clarify whether a given molecular aberration is associated with clinically important effects or not. Nonetheless, determination of genetic polymorphisms, for example, in peripheral blood, is considered an attractive approach for response prediction because this represents a relatively noninvasive approach for molecular profiling. Other methods, for example, immunohistochemistry and RT-PCR based on endoscopically obtained biopsies, always have the inherent doubt of representativity for the entire tumour. On the other hand, a recent study indicated that determination of genetic polymorphisms in peripheral blood may not be representative for the situation in tumour tissue. Thus, Uchida et al (2004) have shown that individuals heterozygous for the 28-bp polymorphism in the TS gene may have cancers that are homozygous for this polymorphism due to loss of one allele during carcinogenesis. Furthermore, they could show that the response to 5-FU-based chemotherapy in these cases was comparable to cases where the entire individual was homozygous. Therefore, it may be superior to analyse the genotype of polymorphisms in tumour cells than in peripheral blood.
In conclusion, our data indicate that determination of the Methylenetetrahydrofolate Reductase C677T polymorphism and the Thymidilate Synthase tandem repeat polymorphism does not provide significant predictive information in multimodally treated oesophageal squamous cell cancer patients. The potential effect of the Methionine Synthase A2756G polymorphism deserves further investigations in similar designed studies.
Change history
16 November 2011
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Ancona E, Ruol A, Santi S, Merigliano S, Sileni VC, Koussis H, Zaninotto G, Bonavina L, Peracchia A (2001) Only pathologic complete response to neoadjuvant chemotherapy improves significantly the long term survival of patients with resectable esophageal squamous cell carcinoma: final report of a randomized, controlled trial of preoperative chemotherapy vs surgery alone. Cancer 91: 2165–2174
Balley LB, Gregory JF (1999) Polymorphism of methylenetetrahydrofolate reductase and other enzymes: metabolic significance, risk and impact on folate requirement. J Nutr 129: 919–922
Brücher BLDM, Stein HJ, Zimmermann F, Werner M, Sarbia M, Busch R, Dittler HJ, Molls M, Fink U, Siewert JR (2004) Responders benefit from neoadjuvant RTx/CTx in esophageal squamous cell carcinoma: results of a prospective phase-II trial. Eur J Surg Oncol 30: 963–971
Chen J, Stampfer MJ, Ma J, Selhub J, Malinow MR, Hennekens CH, Hunter DJ (2001) Influence of a methionine synthase (D919G) polymorphism on plasma homocysteine and folate levels and relation to risk of myocardial infarction. Atherosclerosis 154: 667–672
Choi S-W, Kim YL, Weitzel JN, Mason JB (1998) Folate depletion impairs DNA excision repair in the colon of the rat. Gut 43: 93–99
Cohen V, Panet-Raymond V, Sabbaghian N, Morin I, Batist G, Rozen R (2003) Methylenetetrahydrofolate reductase polymorphism in advanced colorectal cancer: a novel genomic predictor of clinical response to fluoropyrimidine-based chemotherapy. Clin Cancer Res 9: 1611–1615
DiPaolo A, Chu E (2004) The role of Thymidylate Synthase as a molecular biomarker. Clin Cancer Res 10: 411–412
Enzinger PC, Mayer RJ (2003) Esophageal cancer. N Engl J Med 349: 2241–2252
Forastiere AA, Heitmiller RF, Kleinberg L (1997) Multimodality therapy for esophageal cancer. Chest 112: 195–200
Goyette P, Sumner JS, Milos R, Duncan AM, Rosenblatt DS, Matthews RG, Rozen R (1994) Human Methylene tetra Hydro folate Reductase: Isolation of cDNA, Mapping, and Mutation Identification. Nat Genet 7: 551–554
Horie N, Aiba H, Ogaro K, Hojo H, Takeishi K (1995) Functional analysis and DNA polymorphism of the tandemly repeated sequences in the 5′-terminal regulatory region of the human gene for thymidylate synthase. Cell Struct Funct 20: 191–197
Iacopetta B, Grieu F, Joseph D, Elsaleh H (2001) A polymorphism in the enhancer region of the thymidylate synthase promoter influences the survival of colorectal cancer patients treated with 5-fluorouracil. Br J Cancer 85: 827–830
Leclerc D, Campeau E, Goyette P, Adjalla CE, Christensen B, Ross M, Eydoux P, Rosenblatt DS, Rozen R, Gravel RA (1996) Human methionine synthase: cDNA cloning and identification of mutations in patients of the cblG complementation group of folate/cobalamin disorders. Hum Mol Genet 5: 1867–1874
Lerut T, Coosemans W, Leyn PD, Van Raemdonck D, Deneffe G, Decker G (1999) Treatment of esophageal carcinoma. Chest 116: 463–465
Sarbia M, Geddert H, Baldus S, Kiel S, Kandemir Y, Schulz WA, Vossen S, Zotz R, Willers R, Gabbert HE (2005) Methylenetetrahydrofolate Reductase C677T polymorphism and risk of adenocarcinoma of the upper gastrointestinal tract. Scan J Gastroenterol 40: 109–111
Silaste ML, Rantala M, Sampi M, Alfthan G, Aro A, Kesaniemi YA (2001) Polymorphisms of key enzymes in homocysteine metabolism affect diet responsiveness of plasma homocysteine in healthy women. J Nutr 131: 2643–2647
Sobin LH, Wittekind C (1997) TNM Classification of Malignant Tumours 5th edn, Berlin; Springer
Sohn KJ, Croxford R, Yates Z, Lucock M, Kim YI (2004) Effect of the methylenetetrahydrofolate reductase C677T polymorphism on chemosensitivity of colon and breast cancer cells to 5-fluorouracil and methotrexate. J Natl Cancer Inst 96: 134–144
Stahl M, Stuschke M, Lehmann N, Meyer HJ, Walz MK, Seeber S, Klump B, Budach W, Teichmann R, Schmitt M, Schmitt G, Franke C, Wilke H (2005) Chemoradiation with and without surgery in patients with locally advanced squamous cell carcinoma of the esophagus. J Clin Oncol 23: 2310–2317
Stahl M, Wilke H, Fink U, Stuschke M, Walz MK, Siewert JR, Molls M, Fett W, Makoski HB, Breuer N, Schmidt U, Niebel W, Sack H, Eigler FW, Seeber S (1996) Combined preoperative chemotherapy and radiotherapy in patients with locally advanced esophageal cancer: interim analysis of a phase II trial. J Clin Oncol 14: 829–837
Tsai MY, Bignell M, Yang F, Welge BG, Graham KJ, Hanson NQ (2000) Polygenic influence on plasma homocysteine: association of two prevalent mutations, the 844ins68 of cystathionine beta-synthase and A(2756)G of methionine synthase, with lowered plasma homocysteine levels. Atherosclerosis 149: 131–137
Uchida K, Hayashi K, Kawakami K, Schneider S, Yochim JM, Kuramochi H, Takasaki K, Danenberg KD, Danenberg PV (2004) Loss of heterozygosity at the thymidylate synthase (TS) locus on chromosome 18 affects tumor response and survival in individuals heterozygous for a 28-bp polymorphism in the TS gene. Clin Cancer Res 10: 433–439
Villafranca E, Okruzhnov Y, Dominguez MA, Garcia-Foncillas J, Azinovic I, Martinez E, Illarramendi JJ, Arias F, Martinez Monge R, Salgado E, Angeletti S, Brugarolas A (2001) Polymorphisms of the repeated sequences in the enhancer region of the thymidylate synthase gene promoter may predict downstaging after preoperative chemoradiation in rectal cancer. J Clin Oncol 19: 1779–1786
Acknowledgements
Mrs A Jahn, Mrs H Huß, Mrs C Pawlik and Mrs A Haas are gratefully acknowledged for perfect technical assistance. This work was supported by the Deutsche Krebshilfe, Grant Nr. 70-2964 and by the Wilhelm Sander-Stiftung, Grant Nr. 2002.097.1.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Sarbia, M., Stahl, M., von Weyhern, C. et al. The prognostic significance of genetic polymorphisms (Methylenetetrahydrofolate Reductase C677T, Methionine Synthase A2756G, Thymidilate Synthase tandem repeat polymorphism) in multimodally treated oesophageal squamous cell carcinoma. Br J Cancer 94, 203–207 (2006). https://doi.org/10.1038/sj.bjc.6602900
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.bjc.6602900
Keywords
This article is cited by
-
The effects of genomic polymorphisms in one-carbon metabolism pathways on survival of gastric cancer patients received fluorouracil-based adjuvant therapy
Scientific Reports (2016)
-
Germline and somatic genetic predictors of pathological response in neoadjuvant settings of rectal and esophageal cancers: systematic review and meta-analysis
The Pharmacogenomics Journal (2016)
-
A retrospective comparative exploratory study on two Methylentetrahydrofolate Reductase (MTHFR) polymorphisms in esophagogastric cancer: the A1298C MTHFR polymorphism is an independent prognostic factor only in neoadjuvantly treated gastric cancer patients
BMC Cancer (2014)
-
Impact of the MTHFR C677T polymorphism on colorectal cancer in a population with low genetic variability
International Journal of Colorectal Disease (2013)
-
Diet folate, DNA methylation and genetic polymorphisms of MTHFRC677T in association with the prognosis of esophageal squamous cell carcinoma
BMC Cancer (2011)