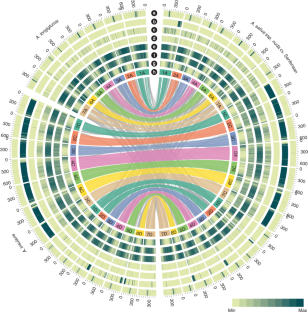
Assemblies of hexaploid cultivated oat, and of close relatives of its diploid and tetraploid progenitors, have revealed its polyploid formation and subgenome evolution. These high-quality oat reference genomes will facilitate the discovery of candidate genes that underlie beneficial traits such as hulless grain and disease resistance.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
References
Van de Peer, Y. et al. The evolutionary significance of polyploidy. Nat. Rev. Genet. 18, 411–424 (2017). This Review correlates polyploidization with environmental change or stress and its short-term adaptive potential.
Yu, H. et al. A route to de novo domestication of wild allotetraploid rice. Cell 184, 1156–1170 (2021). This paper reports a strategy for the polyploidization of diploid rice that can be developed into a staple cereal to strengthen world food security.
Appels, R. et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, eaar7191 (2018). This is a report of an annotated reference sequence representing the hexaploid bread wheat genome, which improved the understanding of wheat biology and genomics-assisted breeding.
Chaffin, A. S. et al. A consensus map in cultivated hexaploid oat reveals conserved grass synteny with substantial subgenome rearrangement. Plant Genome 9, 2 (2016). This paper reports the rearrangement of the hexaploid oat genome relative to its ancestral diploid genomes as a result of frequent translocations among chromosomes.
Miga, K. H. et al. Telomere-to-telomere assembly of a complete human X chromosome. Nature 585, 79–84 (2020). This paper reports the telomere-to-telomere assembly of a human chromosome, which was enabled by ultra-long-read nanopore sequencing.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This is a summary of: Peng, Y. et al. Reference genome assemblies reveal the origin and evolution of allohexaploid oat. Nat. Genet. https://doi.org/10.1038/s41588-022-01127-7 (2022).
Rights and permissions
About this article
Cite this article
Genome origin and evolution of common oat. Nat Genet 54, 1074–1075 (2022). https://doi.org/10.1038/s41588-022-01133-9
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-022-01133-9
