Abstract
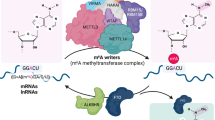
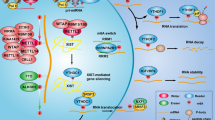
N6-Methyladenosine (m6A) is one of the most abundant modifications of the epitranscriptome and is found in cellular RNAs across all kingdoms of life. Advances in detection and mapping methods have improved our understanding of the effects of m6A on mRNA fate and ribosomal RNA function, and have uncovered novel functional roles in virtually every species of RNA. In this Review, we explore the latest studies revealing roles for m6A-modified RNAs in chromatin architecture, transcriptional regulation and genome stability. We also summarize m6A functions in biological processes such as stem-cell renewal and differentiation, brain function, immunity and cancer progression.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
27 October 2022
In the version of this article initially published, abbreviations but not full names were listed for the adenosines in the Box 1 figure, which have now been restored in the HTML and PDF versions of the article.
References
Wei, C. M., Gershowitz, A. & Moss, B. Methylated nucleotides block 5′ terminus of HeLa cell messenger RNA. Cell 4, 379–386 (1975).
Sommer, S., Lavi, U. & Darnell, J. E. Jr. The absolute frequency of labeled N-6-methyladenosine in HeLa cell messenger RNA decreases with label time. J. Mol. Biol. 124, 487–499 (1978).
Meyer, K. D. et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646 (2012).
Dominissini, D. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012). Together with Meyer et al. (2012), this paper reports the mapping of m6A in the human and mouse transcriptome.
Tegowski, M., Flamand, M. N. & Meyer, K. D. scDART-seq reveals distinct m6A signatures and mRNA methylation heterogeneity in single cells. Mol. Cell 82, 868–878.e10 (2022).
Liu, J. et al. A METTL3–METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95 (2014). This paper reports that the METTL3–METTL14–WTAP complex mediates N6-adenosine methylation.
Wei, C. M. & Moss, B. Nucleotide sequences at the N6-methyladenosine sites of HeLa cell messenger ribonucleic acid. Biochemistry 16, 1672–1676 (1977).
Linder, B. et al. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 12, 767–772 (2015).
Zheng, G. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29 (2013). This study identifies and characterizes ALKBH5 as an m6A demethylase, demonstrating that m6A is a dynamic reversible modification in mRNA.
Jia, G. et al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887 (2011). This study identifies and characterizes FTO as an m6A demethylase, suggesting that m6A is a dynamic reversible modification in mRNA; it should be read in conjunction with Mauer et al. (2017), which suggests that FTO is an m6Am demethylase.
Piekna-Przybylska, D., Decatur, W. A. & Fournier, M. J. The 3D rRNA modification maps database: with interactive tools for ribosome analysis. Nucleic Acids Res. 36, D178–D183 (2008).
Sergiev, P. V., Aleksashin, N. A., Chugunova, A. A., Polikanov, Y. S. & Dontsova, O. A. Structural and evolutionary insights into ribosomal RNA methylation. Nat. Chem. Biol. 14, 226–235 (2018).
Ma, H. et al. N6-Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat. Chem. Biol. 15, 88–94 (2019).
Ren, W. et al. Structure and regulation of ZCCHC4 in m6A-methylation of 28S rRNA. Nat. Commun. 10, 5042 (2019).
Pinto, R. et al. The human methyltransferase ZCCHC4 catalyses N6-methyladenosine modification of 28S ribosomal RNA. Nucleic Acids Res. 48, 830–846 (2020).
van Tran, N. et al. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 47, 7719–7733 (2019).
Liberman, N. et al. N6-Adenosine methylation of ribosomal RNA affects lipid oxidation and stress resistance. Sci. Adv. 6, eaaz4370 (2020).
Xing, M. et al. The 18S rRNA m6A methyltransferase METTL5 promotes mouse embryonic stem cell differentiation. EMBO Rep. 21, e49863 (2020).
Rong, B. et al. Ribosome 18S m6A methyltransferase METTL5 promotes translation initiation and breast cancer cell growth. Cell Rep. 33, 108544 (2020).
Leismann, J. et al. The 18S ribosomal RNA m6A methyltransferase Mettl5 is required for normal walking behavior in Drosophila. EMBO Rep. 21, e49443 (2020).
Ignatova, V. V. et al. The rRNA m6A methyltransferase METTL5 is involved in pluripotency and developmental programs. Genes Dev. 34, 715–729 (2020).
Sendinc, E., Valle-Garcia, D., Jiao, A. & Shi, Y. Analysis of m6A RNA methylation in Caenorhabditis elegans. Cell Discov. 6, 47 (2020).
Sepich-Poore, C. et al. The METTL5–TRMT112 N6-methyladenosine methyltransferase complex regulates mRNA translation via 18S rRNA methylation. J. Biol. Chem. 298, 101590 (2022).
Kierzek, E. & Kierzek, R. The thermodynamic stability of RNA duplexes and hairpins containing N6-alkyladenosines and 2-methylthio-N6-alkyladenosines. Nucleic Acids Res. 31, 4472–4480 (2003).
Meyer, K. D. & Jaffrey, S. R. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. 15, 313–326 (2014).
Liu, N. et al. N6-Methyladenosine-dependent RNA structural switches regulate RNA–protein interactions. Nature 518, 560–564 (2015). This paper describes a mechanism by which m6A affects RNA folding to indirectly have an impact on the binding of proteins to RNA.
Wang, S. et al. The m6A methylation perturbs the Hoogsteen pairing-guided incorporation of an oxidized nucleotide. Chem. Sci. 8, 6380–6388 (2017).
Ashraf, S., Huang, L. & Lilley, D. M. J. Effect of methylation of adenine N6 on kink turn structure depends on location. RNA Biol. 16, 1377–1385 (2019).
Ogawa, A. et al. N6-Methyladenosine (m6A) is an endogenous A3 adenosine receptor ligand. Mol. Cell 81, 659–674.e7 (2021).
Patil, D. P., Pickering, B. F. & Jaffrey, S. R. Reading m6A in the transcriptome: m6A-binding proteins. Trends Cell Biol. 28, 113–127 (2018).
Shi, H., Wei, J. & He, C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell 74, 640–650 (2019).
Zaccara, S., Ries, R. J. & Jaffrey, S. R. Reading, writing and erasing mRNA methylation. Nat. Rev. 20, 608–624 (2019).
Huang, H. et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20, 285–295 (2018).
Edupuganti, R. R. et al. N6-Methyladenosine (m6A) recruits and repels proteins to regulate mRNA homeostasis. Nat. Struct. Mol. Biol. 24, 870–878 (2017).
Ping, X. L. et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24, 177–189 (2014).
Fustin, J. M. et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 155, 793–806 (2013).
Meyer, K. D. et al. 5′ UTR m6A promotes cap-independent translation. Cell 163, 999–1010 (2015).
Wang, X. et al. N6-Methyladenosine modulates messenger RNA translation efficiency. Cell 161, 1388–1399 (2015). Together with Meyer et al. (2015), this paper establishes a role for m6A in regulating mRNA translation.
Zhou, J. et al. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 526, 591–594 (2015).
Li, A. et al. Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res. 27, 444–447 (2017).
Wang, X. et al. N6-Methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120 (2014). This paper establishes a role for m6A in modulating mRNA stability.
Viegas, I. J. et al. N6-Methyladenosine in poly(A) tails stabilize VSG transcripts. Nature 604, 362–370 (2022).
Alarcon, C. R., Lee, H., Goodarzi, H., Halberg, N. & Tavazoie, S. F. N6-Methyladenosine marks primary microRNAs for processing. Nature 519, 482–485 (2015). This paper establishes a role for m6A in modulating miRNA processing.
Alarcon, C. R. et al. HNRNPA2B1 is a mediator of m6A-dependent nuclear RNA processing events. Cell 162, 1299–1308 (2015).
Lence, T. et al. m6A modulates neuronal functions and sex determination in Drosophila. Nature 540, 242–247 (2016).
Haussmann, I. U. et al. m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature 540, 301–304 (2016).
Xiao, W. et al. Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol. Cell 61, 507–519 (2016). This study establishes a role for m6A in modulating splicing.
Roundtree, I. A. et al. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. eLife 6, e31311 (2017).
Edens, B. M. et al. FMRP modulates neural differentiation through m6A-dependent mRNA nuclear export. Cell Rep. 28, 845–854.e5 (2019).
Hsu, P. J. et al. The RNA-binding protein FMRP facilitates the nuclear export of N6-methyladenosine-containing mRNAs. J. Biol. Chem. 294, 19889–19895 (2019).
Wang, Y. et al. N6-Methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 16, 191–198 (2014). This study establishes a role for METTL3 and METTL14 in stemcell renewal via reduced stability of m6A-modified transcripts.
Du, H. et al. YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4–NOT deadenylase complex. Nat. Commun. 7, 12626 (2016).
Park, O. H. et al. Endoribonucleolytic cleavage of m6A-containing RNAs by RNase P/MRP complex. Mol. Cell 74, 494–507.e8 (2019).
Shi, H. et al. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 27, 315–328 (2017).
Choi, J. et al. N6-Methyladenosine in mRNA disrupts tRNA selection and translation-elongation dynamics. Nat. Struct. Mol. Biol. 23, 110–115 (2016).
Taoka, M. et al. Landscape of the complete RNA chemical modifications in the human 80S ribosome. Nucleic Acids Res. 46, 9289–9298 (2018).
Chen, T. et al. m6A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell 16, 289–301 (2015).
Das Mandal, S. & Ray, P. S. Transcriptome-wide analysis reveals spatial correlation between N6-methyladenosine and binding sites of microRNAs and RNA-binding proteins. Genomics 113, 205–216 (2021).
Kanoria, S., Rennie, W. A., Carmack, C. S., Lu, J. & Ding, Y. N6-Methyladenosine enhances post-transcriptional gene regulation by microRNAs. Bioinform Adv. 2, vbab046 (2022).
Chen, L. L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. 21, 475–490 (2020).
Memczak, S. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338 (2013).
Hansen, T. B. et al. Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388 (2013).
Yang, Y. et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 27, 626–641 (2017).
Di Timoteo, G. et al. Modulation of circRNA metabolism by m6A modification. Cell Rep. 31, 107641 (2020).
Pendleton, K. E. et al. The U6 snRNA m6A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell 169, 824–835.e14 (2017). This study shows that METTL16 m6A methyltransferase regulates SAM synthetase mRNA splicing, modulating SAM levels and, thereby, indirectly regulating the methylation of proteins and nucleic acids.
Warda, A. S. et al. Human METTL16 is a N6-methyladenosine (m6A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 18, 2004–2014 (2017).
Huang, H. et al. Histone H3 trimethylation at lysine 36 guides m6A RNA modification co-transcriptionally. Nature 567, 414–419 (2019).
Li, Y. et al. N6-Methyladenosine co-transcriptionally directs the demethylation of histone H3K9me2. Nat. Genet. 52, 870–877 (2020).
Akhtar, J. et al. m6A RNA methylation regulates promoter-proximal pausing of RNA polymerase II. Mol. Cell 81, 3356–3367.e6 (2021).
Xu, W. et al. Dynamic control of chromatin-associated m6A methylation regulates nascent RNA synthesis. Mol. Cell 82, 1156–1168.e7 (2022).
Mendel, M. et al. Methylation of structured RNA by the m6A writer METTL16 is essential for mouse embryonic development. Mol. Cell 71, 986–1000.e11 (2018).
Mendel, M. et al. Splice site m6A methylation prevents binding of U2AF35 to inhibit RNA splicing. Cell 184, 3125–3142.e25 (2021).
Yang, L., Froberg, J. E. & Lee, J. T. Long noncoding RNAs: fresh perspectives into the RNA world. Trends Biochem. Sci. 39, 35–43 (2014).
Patil, D. P. et al. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537, 369–373 (2016). This paper establishes a role for m6A in Xist-mediated silencing.
Nesterova, T. B. et al. Systematic allelic analysis defines the interplay of key pathways in X chromosome inactivation. Nat. Commun. 10, 3129 (2019).
Kopp, F. & Mendell, J. T. Functional classification and experimental dissection of long noncoding RNAs. Cell 172, 393–407 (2018).
Wang, X. et al. N6-Methyladenosine modification of MALAT1 promotes metastasis via reshaping nuclear speckles. Dev. Cell 56, 702–715.e8 (2021).
Andergassen, D. & Rinn, J. L. From genotype to phenotype: genetics of mammalian long non-coding RNAs in vivo. Nat. Rev. Genet. 23, 229–243 (2022).
Liu, J. et al. N6-Methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science 367, 580–586 (2020). This study performs targeted epigenomic editing to test the direct role of m6A on specific RNAs.
Lee, J. H. et al. Enhancer RNA m6A methylation facilitates transcriptional condensate formation and gene activation. Mol. Cell 81, 3368–3385.e9 (2021).
Romanish, M. T., Cohen, C. J. & Mager, D. L. Potential mechanisms of endogenous retroviral-mediated genomic instability in human cancer. Semin. Cancer Biol. 20, 246–253 (2010).
Chelmicki, T. et al. m6A RNA methylation regulates the fate of endogenous retroviruses. Nature 591, 312–316 (2021). This study establishes a role for m6A in suppressing ERVs to help maintain genome stability.
Xu, W. et al. METTL3 regulates heterochromatin in mouse embryonic stem cells. Nature 591, 317–321 (2021).
Niehrs, C. & Luke, B. Regulatory R-loops as facilitators of gene expression and genome stability. Nat. Rev. 21, 167–178 (2020).
Abakir, A. et al. N6-Methyladenosine regulates the stability of RNA:DNA hybrids in human cells. Nat. Genet. 52, 48–55 (2020). This paper establishes a role for m6A in regulating R-loop formation.
Yang, X. et al. m6A promotes R-loop formation to facilitate transcription termination. Cell Res. 29, 1035–1038 (2019).
Zhang, C. et al. METTL3 and N6-methyladenosine promote homologous recombination-mediated repair of DSBs by modulating DNA–RNA hybrid accumulation. Mol. Cell 79, 425–442.e7 (2020).
Geula, S. et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science 347, 1002–1006 (2015). This paper establishes a role for METTL3 in mES cell renewal and differentiation and shows that m6A reduces pluripotency factor transcript stability to facilitate ES cell differentiation.
Meng, T. G. et al. Mettl14 is required for mouse postimplantation development by facilitating epiblast maturation. FASEB J. 33, 1179–1187 (2019).
Zhong, S. et al. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant. Cell 20, 1278–1288 (2008).
Batista, P. J. et al. m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 15, 707–719 (2014).
Aguilo, F. et al. Coordination of m6A mRNA methylation and gene transcription by ZFP217 regulates pluripotency and reprogramming. Cell Stem Cell 17, 689–704 (2015).
Wen, J. et al. Zc3h13 regulates nuclear RNA m6A methylation and mouse embryonic stem cell self-renewal. Mol. Cell 69, 1028–1038.e6 (2018).
Bertero, A. et al. The SMAD2/3 interactome reveals that TGFβ controls m6A mRNA methylation in pluripotency. Nature 555, 256–259 (2018).
Heck, A. M., Russo, J., Wilusz, J., Nishimura, E. O. & Wilusz, C. J. YTHDF2 destabilizes m6A-modified neural-specific RNAs to restrain differentiation in induced pluripotent stem cells. RNA 26, 739–755 (2020).
Zaccara, S. & Jaffrey, S. R. A unified model for the function of YTHDF proteins in regulating m6A-modified mRNA. Cell 181, 1582–1595.e18 (2020).
Yartseva, V. & Giraldez, A. J. The maternal-to-zygotic transition during vertebrate development: a model for reprogramming. Curr. Top. Dev. Biol. 113, 191–232 (2015).
Zhao, B. S. et al. m6A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature 542, 475–478 (2017). This study establishes a role for m6A in maternal transcript clearance during zebrafish MZT.
Kontur, C., Jeong, M., Cifuentes, D. & Giraldez, A. J. Ythdf m6A readers function redundantly during zebrafish development. Cell Rep. 33, 108598 (2020).
Wojtas, M. N. et al. Regulation of m6A transcripts by the 3′ → 5′ RNA helicase YTHDC2 is essential for a successful meiotic program in the mammalian germline. Mol. Cell 68, 374–387.e12 (2017).
Hsu, P. J. et al. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 27, 1115–1127 (2017).
Bailey, A. S. et al. The conserved RNA helicase YTHDC2 regulates the transition from proliferation to differentiation in the germline. eLife 6, e26116 (2017).
Ivanova, I. et al. The RNA m6A reader YTHDF2 is essential for the post-transcriptional regulation of the maternal transcriptome and oocyte competence. Mol. Cell 67, 1059–1067.e4 (2017).
Kasowitz, S. D. et al. Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS Genet. 14, e1007412 (2018).
Sui, X. et al. METTL3-mediated m6A is required for murine oocyte maturation and maternal-to-zygotic transition. Cell Cycle 19, 391–404 (2020).
Giraldez, A. J. et al. Zebrafish miR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312, 75–79 (2006).
Chang, H. et al. Terminal uridylyltransferases execute programmed clearance of maternal transcriptome in vertebrate embryos. Mol. Cell 70, 72–82.e7 (2018).
Wu, Y. & Hirschi, K. K. Regulation of hemogenic endothelial cell development and function. Annu. Rev. Physiol. 83, 17–37 (2021).
Zhang, C. et al. m6A modulates haematopoietic stem and progenitor cell specification. Nature 549, 273–276 (2017).
Lv, J. et al. Endothelial-specific m6A modulates mouse hematopoietic stem and progenitor cell development via Notch signaling. Cell Res. 28, 249–252 (2018).
Lee, H. et al. Stage-specific requirement for Mettl3-dependent m6A mRNA methylation during haematopoietic stem cell differentiation. Nat. Cell Biol. 21, 700–709 (2019).
Vu, L. P. et al. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 23, 1369–1376 (2017).
Weng, H. et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m6A modification. Cell Stem Cell 22, 191–205.e9 (2018).
Yoon, K. J. et al. Temporal control of mammalian cortical neurogenesis by m6A methylation. Cell 171, 877–889.e17 (2017).
Wang, Y. et al. N6-Methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nat. Neurosci. 21, 195–206 (2018).
Li, M. et al. Ythdf2-mediated m6A mRNA clearance modulates neural development in mice. Genome Biol. 19, 69 (2018).
Li, L. et al. Fat mass and obesity-associated (FTO) protein regulates adult neurogenesis. Hum. Mol. Genet. 26, 2398–2411 (2017).
Liu, J., Harada, B. T. & He, C. Regulation of gene expression by N6-methyladenosine in cancer. Trends Cell Biol. 29, 487–499 (2019).
Yang, C. et al. The role of m6A modification in physiology and disease. Cell Death Dis. 11, 960 (2020).
Buffington, S. A., Huang, W. & Costa-Mattioli, M. Translational control in synaptic plasticity and cognitive dysfunction. Annu. Rev. Neurosci. 37, 17–38 (2014).
Merkurjev, D. et al. Synaptic N6-methyladenosine (m6A) epitranscriptome reveals functional partitioning of localized transcripts. Nat. Neurosci. 21, 1004–1014 (2018). This paper reports the identification of subcellular synapse-specific m6A-methylated transcripts.
Yu, J. et al. Dynamic m6A modification regulates local translation of mRNA in axons. Nucleic Acids Res. 46, 1412–1423 (2018).
Widagdo, J. et al. Experience-dependent accumulation of N6-methyladenosine in the prefrontal cortex is associated with memory processes in mice. J. Neurosci. 36, 6771–6777 (2016).
Walters, B. J. et al. The role of the RNA demethylase FTO (fat mass and obesity-associated) and mRNA methylation in hippocampal memory formation. Neuropsychopharmacology 42, 1502–1510 (2017).
Engel, M. et al. The role of m6A/m-RNA methylation in stress response regulation. Neuron 99, 389–403.e9 (2018).
Mauer, J. et al. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature 541, 371–375 (2017). This study characterizes FTO as an m6Am demethylase that regulates mRNA stability, suggesting that m6Am is a dynamic reversable modification; it should be read in conjunction with Jia et al. (2011), which suggests that FTO is a m6A demethylase.
Zhang, Z. et al. METTL3-mediated N6-methyladenosine mRNA modification enhances long-term memory consolidation. Cell Res. 28, 1050–1061 (2018).
Shi, H. et al. m6A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature 563, 249–253 (2018). This study reveals a regulatory role for m6A in neurobehavioural phenotypes by promoting translation of selected transcripts.
Li, H. B. et al. m6A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature 548, 338–342 (2017).
Winkler, R. et al. m6A modification controls the innate immune response to infection by targeting type I interferons. Nat. Immunol. 20, 173–182 (2019).
Liu, Y. et al. N6-Methyladenosine RNA modification-mediated cellular metabolism rewiring inhibits viral replication. Science 365, 1171–1176 (2019). This paper shows that m6A-marked transcripts allow a levelled response to viral infection.
Chow, K. T., Gale, M. Jr & Loo, Y. M. RIG-I and other RNA sensors in antiviral immunity. Annu. Rev. Immunol. 36, 667–694 (2018).
Lu, M. et al. N6-Methyladenosine modification enables viral RNA to escape recognition by RNA sensor RIG-I. Nat. Microbiol. 5, 584–598 (2020).
Qiu, W. et al. N6-Methyladenosine RNA modification suppresses antiviral innate sensing pathways via reshaping double-stranded RNA. Nat. Commun. 12, 1582 (2021).
Chen, Y. G. et al. N6-Methyladenosine modification controls circular RNA immunity. Mol. Cell 76, 96–109.e9 (2019).
Su, R. et al. R-2HG exhibits anti-tumor activity by targeting FTO/m6A/MYC/CEBPA signaling. Cell 172, 90–105.e23 (2018).
Paris, J. et al. Targeting the RNA m6A reader YTHDF2 selectively compromises cancer stem cells in acute myeloid leukemia. Cell Stem Cell 25, 137–148.e6 (2019).
Cheng, Y. et al. N6-Methyladenosine on mRNA facilitates a phase-separated nuclear body that suppresses myeloid leukemic differentiation. Cancer Cell 39, 958–972.e8 (2021).
Cui, Q. et al. m6A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 18, 2622–2634 (2017).
Zhang, S. et al. m6A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell 31, 591–606.e6 (2017).
Liberman, N. et al. Intergenerational hormesis is regulated by heritable 18S rRNA methylation. Preprint at bioRxiv https://doi.org/10.1101/2021/1109.1127.461965 (2021).
Yankova, E. et al. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature 593, 597–601 (2021). This study identifies small molecules that can effectively inhibit METTL3, raising the possibility of therapeutic options for myeloid leukaemia and other m6A-related pathologies.
Kruse, S. et al. A novel synthesis and detection method for cap-associated adenosine modifications in mouse mRNA. Sci. Rep. 1, 126 (2011).
Yuki, H., Kawasaki, H., Imayuki, A. & Yajima, T. Determination of 6-methyladenine in DNA by high-performance liquid chromatography. J. Chromatogr. 168, 489–494 (1979).
Thuring, K., Schmid, K., Keller, P. & Helm, M. LC–MS analysis of methylated RNA. Methods Mol. Biol. 1562, 3–18 (2017).
Liu, N. et al. Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA 19, 1848–1856 (2013).
Imanishi, M., Tsuji, S., Suda, A. & Futaki, S. Detection of N6-methyladenosine based on the methyl-sensitivity of MazF RNA endonuclease. Chem. Commun. 53, 12930–12933 (2017).
Garcia-Campos, M. A. et al. Deciphering the “m6A code” via antibody-independent quantitative profiling. Cell 178, 731–747.e16 (2019).
Dominissini, D. et al. The dynamic N1-methyladenosine methylome in eukaryotic messenger RNA. Nature 530, 441–446 (2016).
Li, X. et al. Transcriptome-wide mapping reveals reversible and dynamic N1-methyladenosine methylome. Nat. Chem. Biol. 12, 311–316 (2016).
Safra, M. et al. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature 551, 251–255 (2017).
Li, X. et al. Base-resolution mapping reveals distinct m1A methylome in nuclear- and mitochondrial-encoded transcripts. Mol. Cell 68, 993–1005.e9 (2017).
Grozhik, A. V. et al. Antibody cross-reactivity accounts for widespread appearance of m1A in 5′ UTRs. Nat. Commun. 10, 5126 (2019).
Harcourt, E. M., Ehrenschwender, T., Batista, P. J., Chang, H. Y. & Kool, E. T. Identification of a selective polymerase enables detection of N6-methyladenosine in RNA. J. Am. Chem. Soc. 135, 19079–19082 (2013).
Hong, T. et al. Precise antibody-independent m6A identification via 4SedTTP-involved and FTO-assisted strategy at single-nucleotide resolution. J. Am. Chem. Soc. 140, 5886–5889 (2018).
Aschenbrenner, J. et al. Engineering of a DNA polymerase for direct m6A sequencing. Angew. Chem. Int. Ed. 57, 417–421 (2018).
Ke, S. et al. A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes Dev. 29, 2037–2053 (2015).
Molinie, B. et al. m6A-LAIC-seq reveals the census and complexity of the m6A epitranscriptome. Nat. Methods 13, 692–698 (2016).
Bringmann, P. & Luhrmann, R. Antibodies specific for N6-methyladenosine react with intact snRNPs U2 and U4/U6. FEBS Lett. 213, 309–315 (1987).
Dai, Q. et al. NM-seq maps 2′-O-methylation sites in human mRNA with base precision. Nat. Methods 14, 695–698 (2017).
Castellanos-Rubio, A. et al. A novel RT-QPCR-based assay for the relative quantification of residue specific m6A RNA methylation. Sci. Rep. 9, 4220 (2019).
Flusberg, B. A. et al. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat. Methods 7, 461–465 (2010).
Garalde, D. R. et al. Highly parallel direct RNA sequencing on an array of nanopores. Nat. Methods 15, 201–206 (2018).
Chen, Y. et al. Study of the whole genome, methylome and transcriptome of Cordyceps militaris. Sci. Rep. 9, 898 (2019).
Pratanwanich, P. N. et al. Identification of differential RNA modifications from nanopore direct RNA sequencing with xPore. Nat. Biotechnol. 39, 1394–1402 (2021).
Mikutis, S. et al. meCLICK-Seq, a substrate-hijacking and RNA degradation strategy for the study of RNA microsoftethylation. ACS Cent. Sci. 6, 2196–2208 (2020).
Liu, W. et al. Identification of a selective DNA ligase for accurate recognition and ultrasensitive quantification of N6-methyladenosine in RNA at one-nucleotide resolution. Chem. Sci. 9, 3354–3359 (2018).
Xiao, Y. et al. An elongation- and ligation-based qPCR amplification method for the radiolabeling-free detection of locus-specific N6-methyladenosine modification. Angew. Chem. Int. Ed. 57, 15995–16000 (2018).
Bokar, J. A., Shambaugh, M. E., Polayes, D., Matera, A. G. & Rottman, F. M. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 3, 1233–1247 (1997). This paper identifies METTL3 as the catalytic subunit of the complex that mediates N6-adenosine methylation.
Clancy, M. J., Shambaugh, M. E., Timpte, C. S. & Bokar, J. A. Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: a potential mechanism for the activity of the IME4 gene. Nucleic Acids Res. 30, 4509–4518 (2002).
Shima, H. et al. S-Adenosylmethionine synthesis is regulated by selective N6-adenosine methylation and mRNA degradation involving METTL16 and YTHDC1. Cell Rep. 21, 3354–3363 (2017).
Akichika, S. et al. Cap-specific terminal N6-methylation of RNA by an RNA polymerase II-associated methyltransferase. Science 363, eaav0080 (2019).
Sun, H., Zhang, M., Li, K., Bai, D. & Yi, C. Cap-specific, terminal N6-methylation by a mammalian m6Am methyltransferase. Cell Res. 29, 80–82 (2019).
Boulias, K. et al. Identification of the m6Am methyltransferase PCIF1 reveals the location and functions of m6Am in the transcriptome. Mol. Cell 75, 631–643.e8 (2019).
Sendinc, E. et al. PCIF1 catalyzes m6Am mRNA methylation to regulate gene expression. Mol. Cell 75, 620–630.e9 (2019).
Belanger, F., Stepinski, J., Darzynkiewicz, E. & Pelletier, J. Characterization of hMTr1, a human Cap1 2′-O-ribose methyltransferase. J. Biol. Chem. 285, 33037–33044 (2010).
Tollervey, D., Lehtonen, H., Jansen, R., Kern, H. & Hurt, E. C. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell 72, 443–457 (1993).
Kiss-Laszlo, Z., Henry, Y., Bachellerie, J. P., Caizergues-Ferrer, M. & Kiss, T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell 85, 1077–1088 (1996).
Nicoloso, M., Qu, L. H., Michot, B. & Bachellerie, J. P. Intron-encoded, antisense small nucleolar RNAs: the characterization of nine novel species points to their direct role as guides for the 2′-O-ribose methylation of rRNAs. J. Mol. Biol. 260, 178–195 (1996).
Tycowski, K. T., Smith, C. M., Shu, M. D. & Steitz, J. A. A small nucleolar RNA requirement for site-specific ribose methylation of rRNA in Xenopus. Proc. Natl Acad. Sci. USA 93, 14480–14485 (1996).
Juhling, F. et al. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 37, D159–D162 (2009).
Ozanick, S., Krecic, A., Andersland, J. & Anderson, J. T. The bipartite structure of the tRNA m1A58 methyltransferase from S. cerevisiae is conserved in humans. RNA 11, 1281–1290 (2005).
Chujo, T. & Suzuki, T. Trmt61B is a methyltransferase responsible for 1-methyladenosine at position 58 of human mitochondrial tRNAs. RNA 18, 2269–2276 (2012).
Vilardo, E. et al. Functional characterization of the human tRNA methyltransferases TRMT10A and TRMT10B. Nucleic Acids Res. 48, 6157–6169 (2020).
Vilardo, E. et al. A subcomplex of human mitochondrial RNase P is a bifunctional methyltransferase — extensive moonlighting in mitochondrial tRNA biogenesis. Nucleic Acids Res. 40, 11583–11593 (2012).
Lafontaine, D., Delcour, J., Glasser, A. L., Desgres, J. & Vandenhaute, J. The DIM1 gene responsible for the conserved m62Am62A dimethylation in the 3′-terminal loop of 18S rRNA is essential in yeast. J. Mol. Biol. 241, 492–497 (1994).
Suvorov, A. N., van Gemen, B. & van Knippenberg, P. H. Increased kasugamycin sensitivity in Escherichia coli caused by the presence of an inducible erythromycin resistance (erm) gene of Streptococcus pyogenes. Mol. Gen. Genet. 215, 152–155 (1988).
Shen, H., Stoute, J. & Liu, K. F. Structural and catalytic roles of the human 18S rRNA methyltransferases DIMT1 in ribosome assembly and translation. J. Biol. Chem. 295, 12058–12070 (2020).
Chen, H. et al. METTL4 is an snRNA m6Am methyltransferase that regulates RNA splicing. Cell Res. 30, 544–547 (2020).
Goh, Y. T., Koh, C. W. Q., Sim, D. Y., Roca, X. & Goh, W. S. S. METTL4 catalyzes m6Am methylation in U2 snRNA to regulate pre-mRNA splicing. Nucleic Acids Res. 48, 9250–9261 (2020).
Liu, F. et al. ALKBH1-mediated tRNA demethylation regulates translation. Cell 167, 816–828.e16 (2016).
Kawarada, L. et al. ALKBH1 is an RNA dioxygenase responsible for cytoplasmic and mitochondrial tRNA modifications. Nucleic Acids Res. 45, 7401–7415 (2017).
Aas, P. A. et al. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature 421, 859–863 (2003).
Xu, C. et al. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 10, 927–929 (2014).
Theler, D., Dominguez, C., Blatter, M., Boudet, J. & Allain, F. H. Solution structure of the YTH domain in complex with N6-methyladenosine RNA: a reader of methylated RNA. Nucleic Acids Res. 42, 13911–13919 (2014).
Xu, C. et al. Structural basis for the discriminative recognition of N6-methyladenosine RNA by the human YT521-B homology domain family of proteins. J. Biol. Chem. 290, 24902–24913 (2015).
Liu, N. et al. N6-Methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 45, 6051–6063 (2017).
Toh, S. M., Xiong, L., Bae, T. & Mankin, A. S. The methyltransferase YfgB/RlmN is responsible for modification of adenosine 2503 in 23S rRNA. RNA 14, 98–106 (2008).
Furuichi, Y. et al. Methylated, blocked 5 termini in HeLa cell mRNA. Proc. Natl Acad. Sci. USA 72, 1904–1908 (1975).
Oerum, S., Degut, C., Barraud, P. & Tisne, C. m1A post-transcriptional modification in tRNAs. Biomolecules 7, 20 (2017).
Peifer, C. et al. Yeast Rrp8p, a novel methyltransferase responsible for m1A 645 base modification of 25S rRNA. Nucleic Acids Res. 41, 1151–1163 (2013).
Waku, T. et al. NML-mediated rRNA base methylation links ribosomal subunit formation to cell proliferation in a p53-dependent manner. J. Cell Sci. 129, 2382–2393 (2016).
Yokoyama, W. et al. rRNA adenine methylation requires T07A9.8 gene as rram-1 in Caenorhabditis elegans. J. Biochem. 163, 465–474 (2018).
Sharma, S. et al. A single N1-methyladenosine on the large ribosomal subunit rRNA impacts locally its structure and the translation of key metabolic enzymes. Sci. Rep. 8, 11904 (2018).
Schwartz, S. m1A within cytoplasmic mRNAs at single nucleotide resolution: a reconciled transcriptome-wide map. RNA 24, 1427–1436 (2018).
Wei, C., Gershowitz, A. & Moss, B. N6,O2′-Dimethyladenosine a novel methylated ribonucleoside next to the 5′ terminal of animal cell and virus mRNAs. Nature 257, 251–253 (1975).
Keith, J. M., Muthukrishnan, S. & Moss, B. Effect of methylation of the N6 position of the penultimate adenosine of capped mRNA on ribosome binding. J. Biol. Chem. 253, 5039–5041 (1978).
Pandey, R. R. et al. The mammalian Cap-specific m6Am RNA methyltransferase PCIF1 regulates transcript levels in mouse tissues. Cell Rep. 32, 108038 (2020).
Ben-Haim, M. S. et al. Dynamic regulation of N6,2′-O-dimethyladenosine (m6Am) in obesity. Nat. Commun. 12, 7185 (2021).
Tartell, M. A. et al. Methylation of viral mRNA cap structures by PCIF1 attenuates the antiviral activity of interferon-β. Proc. Natl Acad. Sci. USA 118, e2025769118 (2021).
Mauer, J. et al. FTO controls reversible m6Am RNA methylation during snRNA biogenesis. Nat. Chem. Biol. 15, 340–347 (2019).
Zorbas, C. et al. The human 18S rRNA base methyltransferases DIMT1L and WBSCR22-TRMT112 but not rRNA modification are required for ribosome biogenesis. Mol. Biol. Cell 26, 2080–2095 (2015).
Liu, K. et al. Regulation of translation by methylation multiplicity of 18S rRNA. Cell Rep. 34, 108825 (2021).
Acknowledgements
Work from the Greer laboratory is supported by grants from the US National Institutes of Health (NIH) (DP2AG055947 and R01AI151215). The authors apologize for literature omitted owing to space limitations.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Genetics thanks Arne Klungland who co-reviewed with Yanjiao Li, Yungui Yang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Hoogsteen base pairing
-
An alternative base pairing in which the purine is flipped and form different hydrogen bonds with partner bases. For adenines, the second hydrogen bond with the pyrimidine base is formed with N6 rather than N1. These alternative base pairs allow for additional structures beyond double helix including triplexes and quadruplexes.
- miRNA microprocessor complex
-
A protein complex involved in the early stages of processing microRNA (miRNA) and RNA interference in animal cells.
- Spliceosome
-
A large RNA–protein complex that catalyses the removal of introns from nuclear pre-mRNA.
- Long non-coding RNAs
-
(lncRNAs). Non-coding RNAs longer than 200 nucleotides.
- Chromosome-associated regulatory RNAs
-
(carRNAs). Regulatory RNAs associated with the chromatin.
- Endogenous retrovirus RNAs
-
(ERVs). The prevalent endogenous viral elements that are derived from retroviruses that have become integrated into the genome.
- R-loops
-
These RNA:DNA hybrids form three-stranded structures when nascent RNA transcripts hybridize with one strand of the DNA template.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Boulias, K., Greer, E.L. Biological roles of adenine methylation in RNA. Nat Rev Genet 24, 143–160 (2023). https://doi.org/10.1038/s41576-022-00534-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41576-022-00534-0
This article is cited by
-
KIT mutations and expression: current knowledge and new insights for overcoming IM resistance in GIST
Cell Communication and Signaling (2024)
-
Identification of ALYREF in pan cancer as a novel cancer prognostic biomarker and potential regulatory mechanism in gastric cancer
Scientific Reports (2024)
-
Natural antisense transcripts as versatile regulators of gene expression
Nature Reviews Genetics (2024)
-
RNA modifications in physiology and disease: towards clinical applications
Nature Reviews Genetics (2024)
-
RNAi-based drug design: considerations and future directions
Nature Reviews Drug Discovery (2024)