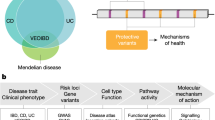
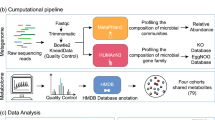
Abstract
Inflammatory bowel disease (IBD) is an immune-mediated disease of the intestinal tract, with complex pathophysiology involving genetic, environmental, microbiome, immunological and potentially other factors. Epidemiological data have provided important insights into risk factors associated with IBD, but are limited by confounding, biases and data quality, especially when pertaining to risk factors in early life. Multiomics platforms provide granular high-throughput data on numerous variables simultaneously and can be leveraged to characterize molecular pathways and risk factors for chronic diseases, such as IBD. Herein, we describe omics platforms that can advance our understanding of IBD risk factors and pathways, and available omics data on IBD and other relevant diseases. We highlight knowledge gaps and emphasize the importance of birth, at-risk and pre-diagnostic cohorts, and neonatal blood spots in omics analyses in IBD. Finally, we discuss network analysis, a powerful bioinformatics tool to assemble high-throughput data and derive clinical relevance.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Torres, J., Mehandru, S., Colombel, J. F. & Peyrin-Biroulet, L. Crohn’s disease. Lancet 389, 1741–1755 (2017).
Ungaro, R., Mehandru, S., Allen, P. B., Peyrin-Biroulet, L. & Colombel, J. F. Ulcerative colitis. Lancet 389, 1756–1770 (2017).
Ananthakrishnan, A. N. et al. Environmental triggers in IBD: a review of progress and evidence. Nat. Rev. Gastroenterol. Hepatol. 15, 39–49 (2018).
Piovani, D. et al. Environmental risk factors for inflammatory bowel diseases: an umbrella review of meta-analyses. Gastroenterology 157, 647–659.e4 (2019).
Agrawal, M. et al. Early life exposures and the risk of inflammatory bowel disease: systematic review and meta-analyses. EClinicalMedicine https://doi.org/10.1016/j.eclinm.2021.100884 (2021).
Schulte, P. A. et al. in Molecular Epidemiology: Principles and Practices, IARC Scientific Publication 163 Ch. 1 (eds Rothman, N. et al.) 1–7 (IARC, 2011).
Torres, J. et al. Results of the seventh scientific workshop of ECCO: precision medicine in IBD– prediction and prevention of inflammatory bowel disease. J. Crohns Colitis 15, 1443–1454 (2021).
Graham, D. B. & Xavier, R. J. Pathway paradigms revealed from the genetics of inflammatory bowel disease. Nature 578, 527–539 (2020).
Rioux, J. D. et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat. Genet. 39, 596–604 (2007).
Ogura, Y. et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature 411, 603–606 (2001).
Liu, J. Z. et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 47, 979–986 (2015).
Inoue, N. et al. Lack of common NOD2 variants in Japanese patients with Crohn’s disease. Gastroenterology 123, 86–91 (2002).
Somineni, H. K. et al. Whole-genome sequencing of African Americans implicates differential genetic architecture in inflammatory bowel disease. Am. J. Hum. Genet. 108, 431–445 (2021).
Spencer, E. A., Helmus, D., Telesco, S., Colombel, J. F. & Dubinsky, M. C. Inflammatory bowel disease clusters within affected sibships in Ashkenazi Jewish multiplex families. Gastroenterology 159, 381–382 (2020).
Gomes, C. F., Jensen, C. B., Allin, K., Torres, J. & Burisch, J. M. Sa509 risk factors associated with familial inflammatory bowel disease [abstract]. Gastroenterology 160, S-528 (2021).
Turpin, W. et al. Associations of NOD2 polymorphisms with Erysipelotrichaceae in stool of in healthy first degree relatives of Crohn’s disease subjects. BMC Med. Genet. 21, 204 (2020).
Miller, G. W. & Jones, D. P. The nature of nurture: refining the definition of the exposome. Toxicol. Sci. 137, 1–2 (2014).
Niedzwiecki, M. M. et al. The exposome: molecules to populations. Annu. Rev. Pharmacol. Toxicol. 59, 107–127 (2019).
Perera, F. P. et al. Effects of transplacental exposure to environmental pollutants on birth outcomes in a multiethnic population. Env. Health Perspect. 111, 201–205 (2003).
Volk, H. E. et al. Prenatal air pollution exposure and neurodevelopment: a review and blueprint for a harmonised approach within ECHO. Environ. Res. 196, 110320 (2021).
Peters, A., Nawrot, T. S. & Baccarelli, A. A. Hallmarks of environmental insults. Cell 184, 1455–1468 (2021).
Agrawal, M., Hillenbrand, C. M., Allin, K. H. & Torres, J. Early life greenspace and the risk of pediatric-onset inflammatory bowel disease: insights into the link between environmental and human health. Gastroenterology 161, 355–357 (2021).
Rundle, A. G. et al. Prenatal exposure to airborne polycyclic aromatic hydrocarbons and childhood growth trajectories from age 5–14 years. Environ. Res. 177, 108595 (2019).
McConnell, R. et al. A longitudinal cohort study of body mass index and childhood exposure to secondhand tobacco smoke and air pollution: the Southern California Children’s Health Study. Env. Health Perspect. 123, 360–366 (2015).
Fleisch, A. F. et al. Prenatal and early life exposure to traffic pollution and cardiometabolic health in childhood. Pediatr. Obes. 12, 48–57 (2017).
Ruokolainen, L. et al. Green areas around homes reduce atopic sensitisation in children. Allergy 70, 195–202 (2015).
Ribeiro, A. I., Tavares, C., Guttentag, A. & Barros, H. Association between neighbourhood green space and biological markers in school-aged children. Findings from the Generation XXI birth cohort. Environ. Int. 132, 105070 (2019).
Elten, M. et al. Ambient air pollution and the risk of pediatric-onset inflammatory bowel disease: a population-based cohort study. Environ. Int. 138, 105676 (2020).
Elten, M. et al. Residential greenspace in childhood reduces risk of pediatric inflammatory bowel disease: a population-based cohort study. Am. J. Gastroenterol. 116, 347–353 (2020).
Lovinsky-Desir, S. et al. Air pollution, urgent asthma medical visits and the modifying effect of neighborhood asthma prevalence. Pediatr. Res. 85, 36–42 (2019).
Mao, G. et al. Individual and joint effects of early-life ambient exposure and maternal prepregnancy obesity on childhood overweight or obesity. Env. Health Perspect. 125, 067005 (2017).
Jedrychowski, W. A. et al. Prenatal exposure to fine particles and polycyclic aromatic hydrocarbons and birth outcomes: a two-pollutant approach. Int. Arch. Occup. Env. Health 90, 255–264 (2017).
Hirten, R. P. et al. Use of physiological data from a wearable device to identify SARS-CoV-2 infection and symptoms and predict COVID-19 diagnosis: observational study. J. Med. Internet Res. 23, e26107 (2021).
Hirten, R. P. et al. Longitudinal autonomic nervous system measures correlate with stress and ulcerative colitis disease activity and predict flare. Inflamm. Bowel Dis. 27, 1576–1584 (2020).
Widbom, L. et al. Elevated plasma cotinine is associated with an increased risk of developing IBD, especially among users of combusted tobacco. PLoS ONE 15, e0235536 (2020).
Walker, D. I. et al. High-resolution metabolomics of occupational exposure to trichloroethylene. Int. J. Epidemiol. 45, 1517–1527 (2016).
Patti, G. J., Yanes, O. & Siuzdak, G. Innovation: Metabolomics: the apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 13, 263–269 (2012).
Li, N. et al. Gestational and childhood exposure to per- and polyfluoroalkyl substances and cardiometabolic risk at age 12 years. Environ. Int. 147, 106344 (2021).
Jedrychowski, W. A. et al. Depressed height gain of children associated with intrauterine exposure to polycyclic aromatic hydrocarbons (PAH) and heavy metals: the cohort prospective study. Environ. Res. 136, 141–147 (2015).
Lochhead, P. et al. Plasma concentrations of perfluoroalkyl substances and risk of inflammatory bowel diseases in women: a nested case control analysis in the Nurses’ Health Study cohorts. Environ. Res. 207, 112222 (2021).
Steenland, K., Zhao, L. & Winquist, A. A cohort incidence study of workers exposed to perfluorooctanoic acid (PFOA). Occup. Env. Med. 72, 373–380 (2015).
Xu, Y. et al. Inflammatory bowel disease and biomarkers of gut inflammation and permeability in a community with high exposure to perfluoroalkyl substances through drinking water. Environ. Res. 181, 108923 (2020).
Canturk, N., Atsu, S. S., Aka, P. S. & Dagalp, R. Neonatal line on fetus and infant teeth: an indicator of live birth and mode of delivery. Early Hum. Dev. 90, 393–397 (2014).
Nair, N. et al. Association between early-life exposures and inflammatory bowel diseases, based on analyses of deciduous teeth. Gastroenterology 159, 383–385 (2020).
Lai, Y. et al. Serum metabolomics identifies altered bioenergetics, signaling cascades in parallel with exposome markers in Crohn’s disease. Molecules 24, 449 (2019).
Kolho, K. L., Pessia, A., Jaakkola, T., de Vos, W. M. & Velagapudi, V. Faecal and serum metabolomics in paediatric inflammatory bowel disease. J. Crohns Colitis 11, 321–334 (2017).
Daniluk, U. et al. Untargeted metabolomics and inflammatory markers profiling in children with Crohn’s disease and ulcerative colitis–a preliminary study. Inflamm. Bowel Dis. 25, 1120–1128 (2019).
Petrick, L. M., Uppal, K. & Funk, W. E. Metabolomics and adductomics of newborn bloodspots to retrospectively assess the early-life exposome. Curr. Opin. Pediatr. 32, 300–307 (2020).
Funk, W. E., Waidyanatha, S., Chaing, S. H. & Rappaport, S. M. Hemoglobin adducts of benzene oxide in neonatal and adult dried blood spots. Cancer Epidemiol. Biomark. Prev. 17, 1896–1901 (2008).
Waidyanatha, S., Zheng, Y., Serdar, B. & Rappaport, S. M. Albumin adducts of naphthalene metabolites as biomarkers of exposure to polycyclic aromatic hydrocarbons. Cancer Epidemiol. Biomark. Prev. 13, 117–124 (2004).
Lydic, T. A. & Goo, Y. H. Lipidomics unveils the complexity of the lipidome in metabolic diseases. Clin. Transl. Med. 7, 4 (2018).
Diab, J. et al. Lipidomics in ulcerative colitis reveal alteration in mucosal lipid composition associated with the disease state. Inflamm. Bowel Dis. 25, 1780–1787 (2019).
Iwatani, S. et al. Novel mass spectrometry-based comprehensive lipidomic analysis of plasma from patients with inflammatory bowel disease. J. Gastroenterol. Hepatol. 35, 1355–1364 (2020).
Murgia, A. et al. Italian cohort of patients affected by inflammatory bowel disease is characterised by variation in glycerophospholipid, free fatty acids and amino acid levels. Metabolomics 14, 140 (2018).
Deyssenroth, M. A. et al. Placental gene networks at the interface between maternal PM2.5 exposure early in gestation and reduced infant birthweight. Environ. Res. 199, 111342 (2021).
Gasparetto, M. et al. Transcription and DNA methylation patterns of blood-derived CD8(+) T cells are associated with age and inflammatory bowel disease but do not predict prognosis. Gastroenterology 160, 232–244.e7 (2021).
Torres, J. et al. Serum biomarkers identify patients who will develop inflammatory bowel diseases up to 5 years before diagnosis. Gastroenterology 159, 96–104 (2020).
Bergemalm, D. et al. Systemic inflammation in pre-clinical ulcerative colitis. Gastroenterology 161, 1526–1539.e9 (2021).
Kim, E. S. et al. Longitudinal changes in fecal calprotectin levels among pregnant women with and without inflammatory bowel disease and their babies. Gastroenterology 160, 1118–1130.e3 (2020).
Ramachandran, N. et al. Self-assembling protein microarrays. Science 305, 86–90 (2004).
Wang, H. et al. Identification of antibody against SNRPB, small nuclear ribonucleoprotein-associated proteins B and B’, as an autoantibody marker in Crohn’s disease using an immunoproteomics approach. J. Crohns Colitis 11, 848–856 (2017).
Kosoy, R. et al. Deep analysis of the peripheral immune system in IBD reveals new insight in disease subtyping and response to monotherapy or combination therapy. Cell. Mol. Gastroenterol. Hepatol. 12, 599–632 (2021).
Verhelst, X. et al. Protein glycosylation as a diagnostic and prognostic marker of chronic inflammatory gastrointestinal and liver diseases. Gastroenterology 158, 95–110 (2020).
Marth, J. D. & Grewal, P. K. Mammalian glycosylation in immunity. Nat. Rev. Immunol. 8, 874–887 (2008).
Miura, Y. et al. BlotGlycoABCTM, an integrated glycoblotting technique for rapid and large scale clinical glycomics. Mol. Cell Proteom. 7, 370–377 (2008).
Dias, A. M. et al. Dysregulation of T cell receptor N-glycosylation: a molecular mechanism involved in ulcerative colitis. Hum. Mol. Genet. 23, 2416–2427 (2014).
Dias, A. M. et al. Metabolic control of T cell immune response through glycans in inflammatory bowel disease. Proc. Natl Acad. Sci. USA 115, e4651–e4660 (2018).
Fujii, H. et al. Core fucosylation on T cells, required for activation of T-cell receptor signaling and induction of colitis in mice, is increased in patients with inflammatory bowel disease. Gastroenterology 150, 1620–1632 (2016).
Trbojević Akmačić, I. et al. Inflammatory bowel disease associates with proinflammatory potential of the immunoglobulin G glycome. Inflamm. Bowel Dis. 21, 1237–1247 (2015).
Clerc, F. et al. Plasma N-glycan signatures are associated with features of inflammatory bowel diseases. Gastroenterology 155, 829–843 (2018).
Bering, S. B. Human milk oligosaccharides to prevent gut dysfunction and necrotizing enterocolitis in preterm neonates. Nutrients 10, 1461 (2018).
Han, S. M., Binia, A., Godfrey, K. M., El-Heis, S. & Cutfield, W. S. Do human milk oligosaccharides protect against infant atopic disorders and food allergy? Nutrients 12, 3212 (2020).
Austin, S. & Bénet, T. Quantitative determination of non-lactose milk oligosaccharides. Anal. Chim. Acta 1010, 86–96 (2018).
Battersby, C., Longford, N., Mandalia, S., Costeloe, K. & Modi, N. Incidence and enteral feed antecedents of severe neonatal necrotising enterocolitis across neonatal networks in England, 2012-13: a whole-population surveillance study. Lancet Gastroenterol. Hepatol. 2, 43–51 (2017).
Autran, C. A. et al. Human milk oligosaccharide composition predicts risk of necrotising enterocolitis in preterm infants. Gut 67, 1064–1070 (2018).
Manthey, C. F., Autran, C. A., Eckmann, L. & Bode, L. Human milk oligosaccharides protect against enteropathogenic Escherichia coli attachment in vitro and EPEC colonisation in suckling mice. J. Pediatr. Gastroenterol. Nutr. 58, 165–168 (2014).
Baron, S. et al. Environmental risk factors in paediatric inflammatory bowel diseases: a population based case control study. Gut 54, 357–363 (2005).
Meda, F., Folci, M., Baccarelli, A. & Selmi, C. The epigenetics of autoimmunity. Cell. Mol. Immunol. 8, 226–236 (2011).
Baccarelli, A. & Bollati, V. Epigenetics and environmental chemicals. Curr. Opin. Pediatr. 21, 243–251 (2009).
Meaburn, E. & Schulz, R. Next generation sequencing in epigenetics: insights and challenges. Semin. Cell Dev. Biol. 23, 192–199 (2012).
Czamara, D. et al. Integrated analysis of environmental and genetic influences on cord blood DNA methylation in newborns. Nat. Commun. 10, 2548 (2019).
Wiklund, P. et al. DNA methylation links prenatal smoking exposure to later life health outcomes in offspring. Clin. Epigenet. 11, 97 (2019).
Deng, Q. et al. The emerging epigenetic role of CD8+ T cells in autoimmune diseases: a systematic review. Front. Immunol. 10, 856 (2019).
Tserel, L. et al. Age-related profiling of DNA methylation in CD8+ T cells reveals changes in immune response and transcriptional regulator genes. Sci. Rep. 5, 13107 (2015).
Ventham, N. T. et al. Integrative epigenome-wide analysis demonstrates that DNA methylation may mediate genetic risk in inflammatory bowel disease. Nat. Commun. 7, 13507 (2016).
Howell, K. J. et al. DNA methylation and transcription patterns in intestinal epithelial cells from pediatric patients with inflammatory bowel diseases differentiate disease subtypes and associate with outcome. Gastroenterology 154, 585–598 (2018).
McDermott, E. et al. DNA methylation profiling in inflammatory bowel disease provides new insights into disease pathogenesis. J. Crohns Colitis 10, 77–86 (2016).
Pittayanon, R. et al. Differences in gut microbiota in patients with vs without inflammatory bowel diseases: a systematic review. Gastroenterology 158, 930–946.e1 (2020).
Torres, J. et al. Infants born to mothers with IBD present with altered gut microbiome that transfers abnormalities of the adaptive immune system to germ-free mice. Gut 69, 42–51 (2020).
Brand, E. C. et al. Healthy cotwins share gut microbiome signatures with their inflammatory bowel disease twins and unrelated patients. Gastroenterology 160, 1970–1985 (2021).
Kurilshikov, A. et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat. Genet. 53, 156–165 (2021).
Galipeau, H. J. et al. Novel fecal biomarkers that precede clinical diagnosis of ulcerative colitis. Gastroenterology 160, 1532–1545 (2021).
Lloyd-Price, J. et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569, 655–662 (2019).
Vich Vila, A. et al. Gut microbiota composition and functional changes in inflammatory bowel disease and irritable bowel syndrome. Sci. Transl. Med. 10, eaap8914 (2018).
Narula, N. et al. Systematic review and meta-analysis: fecal microbiota transplantation for treatment of active ulcerative colitis. Inflamm. Bowel Dis. 23, 1702–1709 (2017).
Iliev, I. D. & Cadwell, K. Effects of intestinal fungi and viruses on immune responses and inflammatory bowel diseases. Gastroenterology 160, 1050–1066 (2021).
Zuo, T. et al. Gut mucosal virome alterations in ulcerative colitis. Gut 68, 1169–1179 (2019).
Leonardi, I. et al. Fungal trans-kingdom dynamics linked to responsiveness to fecal microbiota transplantation (FMT) therapy in ulcerative colitis. Cell Host Microbe 27, 823–829.e3 (2020).
Barr, D. B. et al. The use of dried blood spots for characterising children’s exposure to organic environmental chemicals. Environ. Res. 195, 110796 (2021).
Nørgaard-Pedersen, B. & Simonsen, H. Biological specimen banks in neonatal screening. Acta Paediatr. Suppl. 88, 106–109 (1999).
Nordfalk, F. & Ekstrøm, C. T. Newborn dried blood spot samples in Denmark: the hidden figures of secondary use and research participation. Eur. J. Hum. Genet. 27, 203–210 (2019).
Petrick, L. M. et al. Metabolomics of neonatal blood spots reveal distinct phenotypes of pediatric acute lymphoblastic leukemia and potential effects of early-life nutrition. Cancer Lett. 452, 71–78 (2019).
Thorsen, S. U. et al. Perinatal vitamin D levels are not associated with later risk of developing pediatric-onset inflammatory bowel disease: a Danish case-cohort study. Scand. J. Gastroenterol. 51, 927–933 (2016).
Petralia, F. et al. Integrated proteogenomic characterization across major histological types of pediatric brain cancer. Cell 183, 1962–1985.e31 (2020).
Chen, Y. et al. Variations in DNA elucidate molecular networks that cause disease. Nature 452, 429–435 (2008).
de Souza, H. S. P., Fiocchi, C. & Iliopoulos, D. The IBD interactome: an integrated view of aetiology, pathogenesis and therapy. Nat. Rev. Gastroenterol. Hepatol. 14, 739–749 (2017).
Szklarczyk, D. et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452 (2015).
Petralia, F., Wang, P., Yang, J. & Tu, Z. Integrative random forest for gene regulatory network inference. Bioinformatics 31, i197–i205 (2015).
Peters, L. A. et al. A functional genomics predictive network model identifies regulators of inflammatory bowel disease. Nat. Genet. 49, 1437–1449 (2017).
Khalili, H. et al. Oral contraceptives, reproductive factors and risk of inflammatory bowel disease. Gut 62, 1153–1159 (2013).
Khalili, H. et al. Physical activity and risk of inflammatory bowel disease: prospective study from the Nurses’ Health Study cohorts. BMJ 347, f6633 (2013).
Lo, C. H. et al. Ultra-processed foods and risk of Crohn’s disease and ulcerative colitis: a prospective cohort study. Clin. Gastroenterol. Hepatol. https://doi.org/10.1016/j.cgh.2021.08.031 (2021).
Turpin, W. et al. Analysis of genetic association of intestinal permeability in healthy first-degree relatives of patients with Crohn’s disease. Inflamm. Bowel Dis. 25, 1796–1804 (2019).
Lee, S.-H. et al. Anti-microbial antibody response is associated with future onset of Crohn’s disease independent of biomarkers of altered gut barrier function, subclinical inflammation, and genetic risk. Gastroenterology 161, 1540–1551 (2019).
Acknowledgements
The authors thank J. Gregory, Certified Medical Illustrator, Icahn School of Medicine at Mount Sinai, for the illustrations.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
M.A. is supported by the National Institute of Diabetes and Digestive and Kidney Diseases (K23DK129762-01). J.-F.C. has received research grants from AbbVie, Janssen Pharmaceuticals and Takeda; has received payment for lectures from AbbVie, Amgen, Allergan, Ferring Pharmaceuticals, Shire and Takeda; has received consulting fees from AbbVie, Amgen, Arena Pharmaceuticals, Boehringer Ingelheim, Bristol Myers Squibb, Celgene Corporation, Eli Lilly, Ferring Pharmaceuticals, Galmed Research, Glaxo Smith Kline, Geneva, Iterative Scopes, Janssen Pharmaceuticals, Kaleido Biosciences, Landos, Otsuka, Pfizer, Prometheus, Sanofi, Takeda and TiGenix; and holds stock options in Intestinal Biotech Development. K.H.A., F.P. and T.J. declare no competing interests.
Peer review
Peer review information
Nature Reviews Gastroenterology & Hepatology thanks Anne Griffiths and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Comparative Toxicogenomics Database: http://ctdbase.org/
Human Glycome Project: https://human-glycome.org/
Rights and permissions
About this article
Cite this article
Agrawal, M., Allin, K.H., Petralia, F. et al. Multiomics to elucidate inflammatory bowel disease risk factors and pathways. Nat Rev Gastroenterol Hepatol 19, 399–409 (2022). https://doi.org/10.1038/s41575-022-00593-y
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41575-022-00593-y
This article is cited by
-
Atox1 regulates macrophage polarization in intestinal inflammation via ROS-NLRP3 inflammasome pathway
Journal of Translational Medicine (2024)
-
Targeted delivery of Fc-fused PD-L1 for effective management of acute and chronic colitis
Nature Communications (2024)
-
Mitochondrial function and gastrointestinal diseases
Nature Reviews Gastroenterology & Hepatology (2024)
-
The appendix and ulcerative colitis — an unsolved connection
Nature Reviews Gastroenterology & Hepatology (2023)
-
Inflammation across tissues: can shared cell biology help design smarter trials?
Nature Reviews Rheumatology (2023)