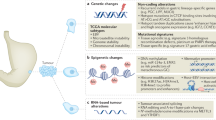
Abstract
Despite improvements in the understanding of cancer causation, much remains unknown regarding the mechanisms by which genomic and non-genomic factors initiate carcinogenesis, drive cell invasion and metastasis, and enable cancer to develop. Technological advances have enabled the analysis of whole genomes, comprising thousands of tumours across populations worldwide, with the aim of identifying mutation signatures associated with particular tumour types. Large collaborative efforts have resulted in the identification and improved understanding of causal factors, and have shed light on new opportunities to prevent cancer. In this new era in cancer genomics, discoveries from studies conducted on an international scale can inform evidence-based strategies in cancer control along the cancer care continuum, from prevention to treatment. In this Review, we present the relevant history and emerging frontiers of cancer genetics and genomics from the perspective of global cancer prevention. We highlight the importance of local context in the adoption of new technologies and emergent evidence, with illustrative examples from worldwide. We emphasize the challenges in implementing important genomic findings in clinical settings with disparate resource availability and present a conceptual framework for the translation of such findings into clinical practice, and evidence-based policies in order to maximize the utility for a population.
Key points
-
Technological advances in germline and tumour genomics are helping to drive progress in elucidating cancer causation at the individual and population level and offer new insights and potential opportunities to prevent certain cancers.
-
The understanding of cancer risks attributable to moderately and highly penetrant mutations in cancer susceptibility genes across different populations has also improved, and genome-wide association studies have led to the identification of single nucleotide polymorphisms that individually confer a small cancer risk and in combination are associated with clinically relevant effects on cancer risk.
-
In practice, taking cancer genomics data to the clinic remains challenging even within some high-income country settings owing to factors including inequitable access and use of limited genetics resources, particularly among minority ethnic and cultural populations; the disproportionate contribution of genomic data from largely European populations also limits the degree to which such information is generalizable to other populations.
-
Optimal strategies to identify individuals at (high) genetic risk for cancer are currently being debated, with evidence increasingly supporting the adoption of population-based genetic testing for individuals with certain cancers.
-
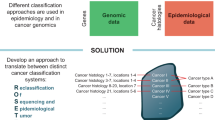
The evidence-to-policy gap in cancer genomics is both deep and wide; the efficiency by which knowledge generation, integration, and dissemination can ultimately lead to better population health can be improved through high-quality implementation research.
-
While genomics can contribute to the pursuit of global health equity through the open exchange of information, expertise and technology between countries of all income levels, this goal will only be possible if it is integrated with the broader, societal understanding of the utility of cancer genomics; an equity lens must be applied throughout the translational research continuum, ensuring adequate representation in research studies of all major world populations, and ethnic and cultural groups.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Huang, K. L. et al. Pathogenic germline variants in 10,389 adult cancers. Cell 173, 355–370.e14 (2018).
Khera, A. V. et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 50, 1219–1224 (2018).
Zhang, Y. D. et al. Assessment of polygenic architecture and risk prediction based on common variants across fourteen cancers. Nat. Commun. 11, 3353 (2020).
Zsofia, K. et al. Genome-wide association studies of cancer. J. Clin. Oncol. 27, 4255–4267 (2010).
McClellan, K. A., Avard, D., Simard, J. & Knoppers, B. M. Personalized medicine and access to health care: potential for inequitable access? Eur. J. Hum. Genet. 21, 143–147 (2013).
National Academies of Sciences, Engineering, and Medicine; Health and Medicine Division; Board on Health Sciences Policy; Roundtable on Genomics and Precision Health. Understanding Disparities in Access to Genomic Medicine: Proceedings of a Workshop (National Academies Press, 2018).
Sirugo, G., Williams, S. M. & Tishkoff, S. A. The missing diversity in human genetic studies. Cell 177, 26–31 (2019).
Yeh, V. M. et al. Can precision medicine actually help people like me? African American and Hispanic perspectives on the benefits and barriers of precision medicine. Ethn. Dis. 30, 149–158 (2020).
Brown, K. F. et al. The fraction of cancer attributable to modifiable risk factors in England, Wales, Scotland, Northern Ireland, and the United Kingdom in 2015. Br. J. Cancer 118, 1130–1141 (2018).
Maniolo, T. A. et al. Finding the missing heritability of complex diseases. Nature 461, 747–753 (2009).
Islami, F. et al. Proportion and number of cancer cases and deaths attributable to potentially modifiable risk factors in the United States. CA Cancer J. Clin. 68, 31–54 (2018).
Brennan, P. & Wild, C. P. Genomics of cancer and a new era for cancer prevention. PLoS Genet. 11, e1005522 (2015).
Tomasetti, C. & Vogelstein, B. Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science 347, 78–81 (2015).
Gotay, C., Dummer, T. & Spinelli, J. Cancer risk: prevention is crucial. Science 347, 728 (2015).
Taubes, G. Epidemiology faces its limits. Science 269, 164–169 (1995).
Higginson, J. Changing concepts in cancer prevention: limitations and implications for future research in environmental carcinogenesis. Cancer Res. 48, 1381–1389 (1988).
Hall, J. M. et al. Linkage of early-onset familial breast cancer to chromosome 17q21. Science 250, 1684–1689 (1990).
Peltomaki, P. et al. Genetic mapping of a locus predisposing to human colorectal cancer. Science 260, 810–812 (1993).
Rahman, N. Realizing the promise of cancer predisposition genes. Nature 505, 302–308 (2014).
Sampson, J. N. et al. Analysis of heritability and shared heritability based on genome-wide association studies for thirteen cancer types. J. Natl Cancer Inst. 107, djv279 (2015).
Weitzel, J. N. et al. Genetics, genomics, and cancer risk assessment: state of the art and future directions in the era of personalized medicine. CA Cancer J. Clin. 61, 327–359 (2011).
Shu, X. et al. Identification of novel breast cancer susceptibility loci in meta-analyses conducted among Asian and European descendants. Nat. Commun. 11, 1217 (2020).
Schumacher, F. R. et al. Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat. Genet. 50, 928–936 (2018).
McKay, J. D. et al. Large-scale association analysis identifies new lung cancer susceptibility loci and heterogeneity in genetic susceptibility across histological subtypes. Nat. Genet. 49, 1126–1132 (2017).
Huyghe, J. R. et al. Discovery of common and rare genetic risk variants for colorectal cancer. Nat. Genet. 51, 76–87 (2019).
Phelan, C. M. et al. Identification of 12 new susceptibility loci for different histotypes of epithelial ovarian cancer. Nat. Genet. 49, 680–691 (2017).
Klein, A. P. et al. Genome-wide meta-analysis identifies five new susceptibility loci for pancreatic cancer. Nat. Commun. 9, 556 (2018).
Scelo, G. et al. Genome-wide association study identifies multiple risk loci for renal cell carcinoma. Nat. Commun. 8, 15724 (2017).
Kachuri, L. et al. Pan-cancer analysis demonstrates that integrating polygenic risk scores with modifiable risk factors improves risk prediction. Preprint at bioRxiv https://doi.org/10.1101/2020.01.28.922088 (2020).
Frank, C. et al. Population landscape of familial cancer. Sci. Rep. 5, 12891 (2015).
Davey Smith, G. & Ebrahim, S. Mendelian randomization: can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 32, 1–22 (2003).
Brennan, P. & Wild, C. Genomics of cancer and a new era for cancer prevention. PLoS Genet. 11, e1005522 (2015).
Loos, R. J. F. & Yeo, G. S. H. The bigger picture of FTO: the first GWAS-identified obesity gene. Nat. Rev. Endocrinol. 10, 51–61 (2014).
Yengo, L. et al. Meta-analysis of genome-wide association studies for height and body mass index in approximately 700000 individuals of European ancestry. Hum. Mol. Genet. 27, 3641–3649 (2018).
World Cancer Research Fund & American Institute for Cancer Research. Body fatness and weight gain and the risk of cancer (WCRF, 2018).
Yu, D. et al. Overall and central obesity and risk of lung cancer: a pooled analysis. J. Natl Cancer Inst. 110, 831–842 (2018).
Mariosa, D. et al. Commentary: what can Mendelian randomization tell us about causes of cancer? Int. J. Epidemiol. 48, 816–821 (2019).
Tsilidis, K. K. et al. Type 2 diabetes and cancer: umbrella review of meta-analyses of observational studies. BMJ 350, g7607 (2015).
Scott, R. A. et al. Common genetic variants highlight the role of insulin resistance and body fat distribution in type 2 diabetes, independent of obesity. Diabetes 63, 4378–4387 (2014).
Johansson, M. et al. The influence of obesity-related factors in the etiology of renal cell carcinoma — a Mendelian randomization study. PLoS Med. 16, e1002724 (2019).
Carreras-Torres, R. et al. The role of obesity, type 2 diabetes, and metabolic factors in pancreatic cancer: a Mendelian randomization study. J. Natl Cancer Inst. 109, djx012 (2017).
Stearns, F. W. One hundred years of pleiotropy: a retrospective. Genetics 186, 767–773 (2010).
Munafò, M. & Davey Smith, G. Repeating experiments is not enough. Nature 553, 400–401 (2018).
Brasky, T. M., White, E. & Chen, C. L. Long-term, supplemental, one-carbon metabolism-related vitamin B use in relation to lung cancer risk in the vitamins and lifestyle (VITAL) cohort. J. Clin. Oncol. 35, 3440–3448 (2017).
Fanidi, A. et al. Is high vitamin B12 status a cause of lung cancer? Int. J. Cancer 145, 1499–1503 (2019).
The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Network. Pan-cancer analysis of whole genomes. Nature 578, 82–93 (2020).
Brash, D. E. et al. A role for sunlight in skin cancer: UV-induced p53 mutations in squamous cell carcinoma. Proc. Natl Acad. Sci. USA 88, 10124–10128 (1991).
Hollstein, M., Sidransky, D., Vogelstein, B. & Harris, C. C. p53 mutations in human cancers. Science 253, 49–53 (1991).
McGregor, W. G. et al. Cell cycle-dependent strand bias for UV-induced mutations in the transcribed strand of excision repair-proficient human fibroblasts but not in repair-deficient cells. Mol. Cell Biol. 11, 1927–1934 (1991).
Denissenko, M. F., Pao, A., Tang, M. & Pfeifer, G. P. Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in P53. Science 274, 430–432 (1996).
Alexandrov, L. B. et al. The repertoire of mutational signatures in human cancer. Nature 578, 94–101 (2020).
Ma, J. et al. The therapeutic significance of mutational signatures from DNA repair deficiency in cancer. Nat. Commun. 9, 3292 (2018).
Ma, X. et al. Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature 555, 371–376 (2018).
Coste, A. et al. Residential exposure to solar ultraviolet radiation and incidence of childhood hematological malignancies in France. Cancer Causes Control 26, 1339–1349 (2015).
Coste, A. et al. Residential exposure to ultraviolet light and risk of precursor B-cell acute lymphoblastic leukemia: assessing the role of individual risk factors, the ESCALE and ESTELLE studies. Cancer Causes Control 10, 1075–1083 (2017).
Scelo, G. et al. Variation in genomic landscape of clear cell renal cell carcinoma across Europe. Nat. Commun. 5, 5135 (2014).
Ng, A. W. T. et al. Aristolochic acids and their derivatives are widely implicated in liver cancers in Taiwan and throughout Asia. Sci. Transl. Med. 9, eaan6446 (2017).
Gerstung, M. et al. The evolutionary history of 2,658 cancers. Nature 578, 122–128 (2020).
Miki, Y. et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266, 66–71 (1994).
Wooster, R. et al. Localization of a breast cancer susceptibility gene, BRCA2, to chromosome 13q12-13. Science 265, 2088–2090 (1994).
McCarthy, A. M. et al. Care segregation, physician recommendation, and racial disparities in BRCA1/2 testing among women with breast cancer. J. Clin. Oncol. 34, 2610–2618 (2016).
Cragun, D. et al. Racial disparities in BRCA testing and cancer risk management across a population-based sample of young breast cancer survivors. Cancer 123, 2497–2505 (2017).
Reid, S. et al. Disparities in BRCA counseling across providers in a diverse population of young breast cancer survivors. Genet. Med. 22, 1088–1093 (2020).
Peterson, J. M. et al. Racial disparities in breast cancer hereditary risk assessment referrals. J. Genet. Couns. 29, 587–593 (2020).
Cruz-Correa, M. et al. Clinical cancer genetics disparities among Latinos. J. Genet. Couns. 26, 379–386 (2017).
Walsh, T. et al. Detection of inherited mutations for breast and ovarian cancer using genomic capture and massively parallel sequencing. Proc. Natl Acad. Sci. USA 107, 12629–12633 (2010).
Graffeo, R. et al. Time to incorporate germline multigene panel testing into breast and ovarian cancer patient care. Breast Cancer Res. Treat. 160, 393–410 (2016).
Gallego, C. J. et al. Next-generation sequencing panels for the diagnosis of colorectal cancer and polyposis syndromes: a cost-effectiveness analysis. J. Clin. Oncol. 33, 2084–2091 (2015).
Li, H. et al. Breast cancer risk prediction using a polygenic risk score in the familial setting: a prospective study from the breast cancer family registry and kConFab. Genet. Med. 19, 30–35 (2017).
Kurian, A. W. et al. Uptake, results, and outcomes of germline multiple-gene sequencing after diagnosis of breast cancer. JAMA Oncol. 4, 1066–1072 (2018).
Asphaug, L. & Melberg, H. O. The cost-effectiveness of multigene panel testing for hereditary breast and ovarian cancer in Norway. MDM Policy Pract. 4, 2381468318821103 (2019).
Manchanda, R. et al. Cost-effectiveness of population-based BRCA1, BRCA2, RAD51C, RAD51D, BRIP1, PALB2 mutation testing in unselected general population women. J. Natl Cancer Inst. 110, 714–725 (2018).
Buys, S. S. et al. A study of over 35,000 women with breast cancer tested with a 25-gene panel of hereditary cancer genes. Cancer 12310, 1721–1730 (2017).
Sun, L. et al. A cost-effectiveness analysis of multigene testing for all patients with breast cancer. JAMA Oncol. 5, 1718–1730 (2019).
Kamps, R. et al. Next-generation sequencing in oncology: genetic diagnosis, risk prediction and cancer classification. Int. J. Mol. Sci. 18, E308 (2017).
Kapoor, N. S. et al. Multigene panel testing detects equal rates of pathogenic BRCA1/2 mutations and has a higher diagnostic yield compared to limited BRCA1/2 analysis alone in patients at risk for hereditary breast cancer. Ann. Surg. Oncol. 22, 3282–3288 (2015).
Crawford, B. et al. Multi-gene panel testing for hereditary cancer predisposition in unsolved high-risk breast and ovarian cancer patients. Breast Cancer Res. Treat. 163, 383–390 (2017).
Roberts, M. E. et al. CDH1 penetrance estimates in clinically ascertained families vs families ascertained for multiple gastric cancers. JAMA Oncol. 5, 1325–1331 (2019).
Hauke, J. et al. Gene panel testing of 5589 BRCA1/2-negative index patients with breast cancer in a routine diagnostic setting: results of the German consortium for hereditary breast and ovarian cancer. Cancer Med. 7, 1349–1358 (2018).
Robson, M. E. et al. American Society of Clinical Oncology policy statement update: genetic and genomic testing for cancer susceptibility. J. Clin. Oncol. 33, 3660–3667 (2015).
Eccles, D. M. et al. BRCA1 and BRCA2 genetic testing — pitfalls and recommendations for managing variants of uncertain clinical significance. Ann. Oncol. 26, 2057–2065 (2015).
Hall, M. J. & Forman, A. D. Gene panel testing for inherited cancer risk. J. Natl Compr. Cancer Netw. 12, 1339–1346 (2014).
Mersch, J. et al. Prevalence of variant reclassification following hereditary cancer genetic testing. JAMA 320, 1266–1274 (2018).
Clain, E. et al. Availability and payer coverage of BRCA1/2 tests and gene panels. Nat. Biotechnol. 33, 900–902 (2015).
Cheng, D. T. et al. Comprehensive detection of germline variants by MSK-IMPACT, a clinical diagnostic platform for solid tumor molecular oncology and concurrent cancer predisposition testing. BMC Med. Genomics 10, 33 (2017).
Henn, J. et al. Diagnostic yield and clinical utility of a comprehensive gene panel for hereditary tumor syndromes. Hered. Cancer Clin. Pract. 17, 5 (2019).
Chen, S. et al. Characterization of BRCA1 and BRCA2 mutations in a large United States sample. J. Clin. Oncol. 24, 863–871 (2006).
Evans, D. G. et al. Penetrance estimates for BRCA1 and BRCA2 based on genetic testing in a clinical cancer genetics service setting: risks of breast/ovarian cancer quoted should reflect the cancer burden in the family. BMC Cancer 8, 155 (2008).
Gabai-Kapara, E. et al. Population-based screening for breast and ovarian cancer risk due to BRCA1 and BRCA2. Proc. Natl Acad. Sci. USA 111 (39), 14205–14210 (2014).
Metcalfe, K. A. et al. The risk of breast cancer in BRCA1 and BRCA2 mutation carriers without a first-degree relative with breast cancer. Clin. Genet. 93, 1063–1068 (2018).
Kuchenbaecker, K. B. et al. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA 317, 2402–2416 (2017).
van der Post, R. S. et al. Hereditary diffuse gastric cancer: updated clinical guidelines with an emphasis on germline CDH1 mutation carriers. J. Med. Genet. 52, 361–374 (2015).
Pharoah, P. D., Guilford, P. & Caldas, C. Incidence of gastric cancer and breast cancer in CDH1 (E-cadherin) mutation carriers from hereditary diffuse gastric cancer families. Gastroenterology 121, 1348–1353 (2001).
Foulkes, W., Knoppers, B. & Turnbull, C. Population genetic testing for cancer susceptibility: founder mutations to genomes. Nat. Rev. Clin. Oncol. 13, 41–54 (2016).
King, M., Levy-Lahad, E. & Lahad, A. Population-based screening for BRCA1 and BRCA2: 2014 Lasker award. JAMA 312, 1091–1092 (2014).
Beitsch, P. D. et al. Underdiagnosis of hereditary breast cancer: are genetic testing guidelines a tool or an obstacle? J. Clin. Oncol. 37, 453–460 (2019).
LaDuca, H. et al. A clinical guide to hereditary cancer panel testing: evaluation of gene-specific cancer associations and sensitivity of genetic testing criteria in a cohort of 165,000 high-risk patients. Genet. Med. 13, 1–9 (2019).
Childers, C. P., Childers, K. K., Maggard-Gibbons, M. & Macinko, J. National estimates of genetic testing in women with a history of breast or ovarian cancer. J. Clin. Oncol. 35, 3800 (2017).
Nielsen, S. M. et al. Genetic testing and clinical management practices for variants in non-BRCA1/2 breast (and breast/ovarian) cancer susceptibility genes: an international survey by the evidence-based network for the interpretation of germline mutant alleles (ENIGMA) clinical working group. JCO Precis. Oncol. https://doi.org/10.1200/PO.18.00091 (2018).
Kurian, A. W. et al. Genetic testing and results in a population-based cohort of breast cancer patients and ovarian cancer patients. J. Clin. Oncol. 37, 1305–1315 (2019).
Khoury, M. J., McCabe, L. L. & McCabe, E. R. Population screening in the age of genomic medicine. N. Engl. J. Med. 348, 50–58 (2003).
National Comprehensive Cancer Network. Clinical practice guidelines in oncology. Genetic/familial high-risk assessment: breast, ovarian, and pancreatic. NCCN https://www.nccn.org/professionals/physician_gls/pdf/genetics_bop.pdf (2019).
Heemskerk-Gerritsen, B. A. M. et al. Survival after bilateral risk-reducing mastectomy in healthy BRCA1 and BRCA2 mutation carriers. Breast Cancer Res. Treat. 177, 723–733 (2019).
Finch, A. P. et al. Impact of oophorectomy on cancer incidence and mortality in women with a BRCA1 or BRCA2 mutation. J. Clin. Oncol. 32, 1547–1553 (2014).
Domchek, S. M. et al. Association of risk-reducing surgery in BRCA1 or BRCA2 mutation carriers with cancer risk and mortality. JAMA 304, 967–975 (2010).
Yang, S. et al. Underdiagnosis of hereditary breast and ovarian cancer in Medicare patients: genetic testing criteria miss the mark. Ann. Surg. Oncol. 25, 2925–2931 (2018).
Taylor, A. & Tischkowitz, M. Caveat emptor: the perils of panel testing in hereditary breast cancer. J. Clin. Oncol. 37, 2176–2177 (2019).
Manahan, E. R. et al. Consensus guidelines on genetic testing for hereditary breast cancer from the American Society of Breast Surgeons. Ann. Surg. Oncol. 26, 3025–3031 (2019).
Hughes, S. K. A pathway for identifying women at increased risk for breast cancer and providing personalized management and risk reduction. Oncol. Issues 32, 38–47 (2017).
Caswell-Jin., J. L. et al. Cascade genetic testing of relatives for hereditary cancer risk: results of an online initiative. J. Natl Cancer Inst. 111, 95–98 (2019).
MacDonald, D. J. et al. Selection of family members for communication of cancer risk and barriers to this communication before and after genetic cancer risk assessment. Genet. Med. 9, 275–282; erratum 9, 483 (2007).
Forrest, K. et al. To tell or not to tell: barriers and facilitators in family communication about genetic risk. Clin. Genet. 64, 317–326 (2003).
Ricker, C. N. et al. Patient communication of cancer genetic test results in a diverse population. Transl. Behav. Med. 8, 85–94 (2018).
Landsbergen, K., Verhaak, C., Floor Kraaimaat, F. & Hoogerbrugge, N. Genetic uptake in BRCA-mutation families is related to emotional and behavioral communication characteristics of index patients. Familial Cancer 4, 115–119 (2005).
Joseph, L. et al. The spectrum of clinical utilities in molecular pathology testing procedures for inherited conditions and cancer: a report of the association for molecular pathology. J. Mol. Diagn. 18, 605–619 (2016).
Tandy-Connor, S. et al. False-positive results released by direct-to-consumer genetic tests highlight the importance of clinical confirmation testing for appropriate patient care. Genet. Med. 20, 1515–1521 (2018).
Farmer, M. B. et al. Adverse events in genetic testing: the fourth case series. Cancer J. 25, 231–236 (2019).
Hudson, K., Javittm, G., Burke, W. & Byers, P. ASHG statement on direct-to-consumer genetic testing in the United States. Am. J. Hum. Genetics. 81, 635–637 (2007).
Wade, C. H. & Wilfond, B. S. Ethical and clinical practice considerations for genetic counselors related to direct-to-consumer marketing of genetic tests. Am. J. Med. Genet. C Semin. Med. Genet. 142C, 284–292 (2006).
US Preventive Services Task Force. et al. Risk assessment, genetic counseling, and genetic testing for BRCA-related cancer: US preventive services task force recommendation statement. JAMA 322, 652–665 (2019); erratum 322, 1830 (2019).
Biswas, S. et al. A two-stage approach to genetic risk assessment in primary care. Breast Cancer Res. Treat. 155, 375–383 (2016).
Jia, G. et al. Evaluating the utility of polygenic risk scores in identifying high-risk individuals for eight common cancers. JNCI Cancer Spectr. 4, pkaa021 (2020).
Fischer, C. et al. Evaluating the performance of the breast cancer genetic risk models BOADICEA, IBIS, BRCAPRO and Claus for predicting BRCA1/2 mutation carrier probabilities: a study based on 7352 families from the German Hereditary Breast and Ovarian Cancer Consortium. J. Med. Genet. 50, 360–367 (2013).
Ashton-Prolla, P. et al. Development and validation of a simple questionnaire for the identification of hereditary breast cancer in primary care. BMC Cancer 9, 283 (2009).
Bellcross, C. A. et al. Evaluation of a breast/ovarian cancer genetics referral screening tool in a mammography population. Genet. Med. 11, 783–789 (2009).
Hoskins, K. F., Zwaagstra, A. & Ranz, M. Validation of a tool for identifying women at high risk for hereditary breast cancer in population-based screening. Cancer 107, 1769–1776 (2006).
Oros, K. K. et al. Application of BRCA1 and BRCA2 mutation carrier prediction models in breast and/or ovarian cancer families of French Canadian descent. Clin. Genet. 70, 320–329 (2006).
Gilpin, C. A., Carson, N. & Hunter, A. G. A preliminary validation of a family history assessment form to select women at risk for breast or ovarian cancer for referral to a genetics center. Clin. Genet. 58, 299–308 (2000).
Domchek, S. & Robson, M. Broadening criteria for BRCA1/2 evaluation: placing the recommendation in context. JAMA 322, 619–621 (2019).
Terry, M. B. et al. 10-year performance of four models of breast cancer risk: a validation study. Lancet Oncol. 20, 504–517 (2019).
Mavaddat, N. et al. Polygenic risk scores for prediction of breast cancer and breast cancer subtypes. Am. J. Hum. Genet. 104, 21–34 (2019).
Wojcik, G. L. et al. Genetic analyses of diverse populations improves discovery for complex traits. Nature 19, 1 (2019).
Gurdasani, D., Barroso, I., Zeggini, E. & Sandhu, M. S. Genomics of disease risk in globally diverse populations. Nat. Rev. Genet. 20, 520–535 (2019).
Martin, A. R. et al. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 51, 584–591 (2019).
Human Heredity & Health in Africa. H3Africahttps://h3africa.org/index.php/consortium (2020).
Andermann, A., Blancquaert, I., Beauchamp, S. & Déry, V. Revisiting Wilson and Jungner in the genomic age: a review of screening criteria over the past 40 years. Bull. World Health Organ. 86, 317–319 (2008).
Wilson, J. M. & Jungner, Y. G. Principles and practice of mass screening for disease (WHO, 1968).
Levy-Lahad, E. et al. Founder BRCA1 and BRCA2 mutations in Ashkenazi Jews in Israel: frequency and differential penetrance in ovarian cancer and in breast-ovarian cancer families. Am. J. Hum. Genet. 60, 1059–1067 (1997).
Oddoux, C. et al. The carrier frequency of the BRCA2 6174delT mutation among Ashkenazi Jewish individuals is approximately 1%. Nat. Genet. 14, 188 (1996).
Roa, B. B., Boyd, A. A., Volcik, K. & Richards, C. S. Ashkenazi Jewish population frequencies for common mutations in BRCA1 and BRCA2. Nat. Genet. 14, 185 (1996).
Struewing, J. P. et al. The carrier frequency of the BRCA1 185delAG mutation is approximately 1 percent in Ashkenazi Jewish individuals. Nat. Genet. 11, 198 (1995).
Hartge, P. et al. The prevalence of common BRCA1 and BRCA2 mutations among Ashkenazi Jews. Am. J. Hum. Genet. 64, 963–970 (1999).
Frank, T. S. et al. Clinical characteristics of individuals with germline mutations in BRCA1 and BRCA2: analysis of 10,000 individuals. J. Clin. Oncol. 20, 1480–1490 (2002).
Hirsh-Yechezkel, G. et al. Population attributes affecting the prevalence of BRCA mutation carriers in epithelial ovarian cancer cases in Israel. Gynecol. Oncol. 89, 494–498 (2003).
Lu, K. H. et al. A population-based study of BRCA1 and BRCA2 mutations in Jewish women with epithelial ovarian cancer. Obstet. Gynecol. 93, 34–37 (1999).
King, M. C., Marks, J. H. & Mandell, J. B. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science 302, 643–646 (2003).
Warner, E. et al. Prevalence and penetrance of BRCA1 and BRCA2 gene mutations in unselected Ashkenazi Jewish women with breast cancer. J. Natl Cancer Inst. 91, 1241–1247 (1999).
Antoniou, A. et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: a combined analysis of 22 studies. Am. J. Hum. Genet. 72, 1117–1130 (2003).
Satagopan, J. M. et al. Ovarian cancer risk in Ashkenazi Jewish carriers of BRCA1 and BRCA2 mutations. Clin. Cancer Res. 8, 3776–3781 (2002).
Satagopan, J. M. et al. The lifetime risks of breast cancer in Ashkenazi Jewish carriers of BRCA1 and BRCA2 mutations. Cancer Epidemiol. Biomarkers Prev. 10, 467–473 (2001).
Shkedi-Rafid, S. et al. BRCA genetic testing of individuals from families with low prevalence of cancer: experiences of carriers and implications for population screening. Genet. Med. 14, 688–694 (2012).
Metcalfe, K. A. et al. Long-term follow-up of Jewish women with a BRCA1 and BRCA2 mutation who underwent population genetic screening. Breast Cancer Res. Treat. 133, 735–740 (2012).
Lieberman, S. et al. Familial communication and cascade testing among relatives of BRCA population screening participants. Genet. Med. 20, 1446–1454 (2018).
Rubinstein, W. S., Jiang, H., Dellefave, L. & Rademaker, A. W. Cost-effectiveness of population-based BRCA1/2 testing and ovarian cancer prevention for Ashkenazi Jews: a call for dialogue. Genet. Med. 11, 629 (2009).
Domchek, S. M. et al. Mortality after bilateral salpingo-oophorectomy in BRCA1 and BRCA2 mutation carriers: a prospective cohort study. Lancet Oncol. 7, 223–229 (2006).
Rebbeck, T. R. et al. Prophylactic oophorectomy in carriers of BRCA1 or BRCA2 mutations. N. Engl. J. Med. 346, 1616–1622 (2002).
Manchanda, R. et al. Cost-effectiveness of population based BRCA testing with varying Ashkenazi Jewish ancestry. Am. J. Obstet. Gynecol. 217, 578.e1–578.e12 (2017).
De Felice, F. et al. Bilateral risk-reduction mastectomy in BRCA1 and BRCA2 mutation carriers: a meta-analysis. Ann. Surg. Oncol. 22, 2876–2880 (2015).
Eleje, G. U. et al. Risk-reducing bilateral salpingo-oophorectomy in women with BRCA1 or BRCA2 mutations. Cochrane Database Syst. Rev. 8, CD012464 (2018).
Achatz, M. I. & Zambetti, G. P. The inherited p53 mutation in the Brazilian population. Cold Spring Harb. Perspect. Med. 6, a026195 (2016).
Giacomazzi, J. et al. Prevalence of the TP53 p.R337H mutation in breast cancer patients in Brazil. PLoS ONE 9, e99893 (2014).
Custódio, G. et al. Impact of neonatal screening and surveillance for the TP53 R337H mutation on early detection of childhood adrenocortical tumors. J. Clin. Oncol. 31, 2619–2626 (2013).
Robson, M. & Domchek, S. Broad application of multigene panel testing for breast cancer susceptibility — Pandora’s box is opening wider. JAMA Oncol. 5, 1687–1688 (2019).
Ginsburg, O. & Brennan, P. Genetic testing for breast cancer in the era of multigene panels: can we make an impact on population health? J. Clin. Oncol. 36, 2817–2819 (2018).
Zheng, Y. et al. Inherited breast cancer in Nigerian women. J. Clin. Oncol. 36, 2820 (2018).
Fernandes, G. C. et al. Prevalence of BRCA1/BRCA2 mutations in a Brazilian population sample at risk for hereditary breast cancer and characterization of its genetic ancestry. Oncotarget 7, 80465–80481 (2016).
Alemar, B. et al. Prevalence of Hispanic BRCA1 and BRCA2 mutations among hereditary breast and ovarian cancer patients from Brazil reveals differences among Latin American populations. Cancer Genet. 209, 417–422 (2016).
Achatz, M. I. et al. Recommendations for advancing the diagnosis and management of hereditary breast and ovarian cancer in Brazil. JCO Glob. Oncol. 6, 439–452 (2020).
Simoes Correa-Galendi, J., Del Pilar Estevez Diz, M., Stock, S. & Müller, D. Economic modelling of screen-and-treat strategies for Brazilian women at risk of hereditary breast and ovarian cancer. Appl. Health Econ. Health Policy https://doi.org/10.1007/s40258-020-00599-0 (2020).
Ramos, M. C. A. et al. Cost effectiveness of the cancer prevention program for carriers of the BRCA1/2 mutation. Rev. Saude Publica 52, 94 (2018).
Nakamura, S. et al. Current status of the management of hereditary breast and ovarian cancer in Asia: first report by the Asian BRCA consortium. Public Health Genomics 19, 53–60 (2016).
Manolio, T. A. et al. Opportunities, resources, and techniques for implementing genomics in clinical care. Lancet 394, 511–520 (2019).
Bray, F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 (2018).
Bray, F. & Soerjomataram, I. Population attributable fractions continue to unmask the power of prevention. Br. J. Cancer 118, 1031–1032 (2018).
Romero, Y. et al. National cancer control plans: a global analysis. Lancet Oncol. 19, e546–e555 (2018).
Della Valle, A. et al. A snapshot of current genetic testing practice in Lynch syndrome: the results of a representative survey of 33 Latin American existing centres/registries. Eur. J. Cancer 119, 112–121 (2019).
Ashton-Prolla, P. & Seuanez, H. N. The Brazilian Hereditary Cancer Network: historical aspects and challenges for clinical cancer genetics in the public health care system in Brazil. Genet. Mol. Biol. 39, 163–165 (2016).
Kwong, A. et al. Comprehensive spectrum of BRCA1 and BRCA2 deleterious mutations in breast cancer in Asian countries. J. Med. Genet. 53, 15 (2016).
Westfall, J. M., Mole, J. & Fagnan, L. Practice-based research — “Blue Highways” on the NIH roadmap. JAMA 297, 403–406 (2007).
Lenfant, C. Shattuck lecture — clinical research to clinical practice — lost in translation? N. Engl. J. Med. 349, 868–874 (2003).
Schully, S. D. et al. Translational research in cancer genetics: the road less traveled. Public Health Genomics 14, 1–8 (2011).
Trinh-Shevrin, C. et al. Defining an integrative approach for health promotion and disease prevention: a population health equity framework. J. Health Care Poor Underserved 26, 146–163 (2015).
Burke, W. et al. The path from genome-based research to population health: development of an international public health genomics network. Genet. Med. 8, 451–458 (2006).
World Health Organization. Human genomics in global health. WHO https://www.who.int/genomics/en/ (2020).
Ferreira, A. M. et al. Clinical spectrum of Li-Fraumeni syndrome/Li-Fraumeni-like syndrome in Brazilian individuals with the TP53 p.R337H mutation. J. Steroid Biochem. Mol. Biol. 190, 250–255 (2019).
Acknowledgements
The authors acknowledge the contributions of A. Newcomb for assistance with the design of Fig. 2 and A. Li for administrative support.
Author information
Authors and Affiliations
Contributions
O.G. and P.B. contributed equally to the conceptualization and development of this manuscript. All authors made a substantial contribution to all aspects of the preparation of this manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Disclaimer
Where authors are identified as personnel of the International Agency for Research on Cancer/World Health Organization, the authors alone are responsible for the views expressed in this article, and they do not necessarily represent the decisions, policy, or views of the International Agency for Research on Cancer/World Health Organization.
Peer review information
Nature Reviews Clinical Oncology thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
CRUK Mutographs: https://mutographs.org
NCI Sherlock: https://dceg.cancer.gov/research/cancer-types/lung/sherlock-lung-study
Rights and permissions
About this article
Cite this article
Ginsburg, O., Ashton-Prolla, P., Cantor, A. et al. The role of genomics in global cancer prevention. Nat Rev Clin Oncol 18, 116–128 (2021). https://doi.org/10.1038/s41571-020-0428-5
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41571-020-0428-5
This article is cited by
-
Trends in orphan medicinal products approvals in the European Union between 2010–2022
Orphanet Journal of Rare Diseases (2024)
-
Mutational signatures and processes in hepatobiliary cancers
Nature Reviews Gastroenterology & Hepatology (2022)
-
Colorectal cancer organoid models uncover oxaliplatin-resistant mechanisms at single cell resolution
Cellular Oncology (2022)
-
Comparing models of delivery for cancer genetics services among patients receiving primary care who meet criteria for genetic evaluation in two healthcare systems: BRIDGE randomized controlled trial
BMC Health Services Research (2021)