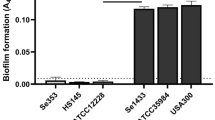
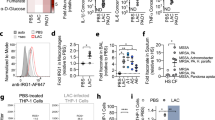
Abstract
Staphylococcus aureus is a leading cause of biofilm-associated prosthetic joint infection (PJI), resulting in considerable disability and prolonged treatment. It is known that host leukocyte IL-10 production is required for S. aureus biofilm persistence in PJI. An S. aureus bursa aurealis Tn library consisting of 1,952 non-essential genes was screened for mutants that failed to induce IL-10 in myeloid-derived suppressor cells (MDSCs), which identified a critical role for bacterial lactic acid biosynthesis. We generated an S. aureus ddh/ldh1/ldh2 triple Tn mutant that cannot produce d- or l-lactate. Co-culture of MDSCs or macrophages with ddh/ldh1/ldh2 mutant biofilm produced substantially less IL-10 compared with wild-type S. aureus, which was also observed in a mouse model of PJI and led to reduced biofilm burden. Using MDSCs recovered from the mouse PJI model and in vitro leukocyte–biofilm co-cultures, we show that bacterial-derived lactate inhibits histone deacetylase 11, causing unchecked HDAC6 activity and increased histone 3 acetylation at the Il-10 promoter, resulting in enhanced Il-10 transcription in MDSCs and macrophages. Finally, we show that synovial fluid of patients with PJI contains elevated amounts of d-lactate and IL-10 compared with control subjects, and bacterial lactate increases IL-10 production by human monocyte-derived macrophages.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The ChIP–seq and RNA-seq datasets are available in the GEO repository (accession number GSE135496). Source data are provided with this paper.
Code availability
All codes used are published programs, with citations for each provided in the references.
References
Percival, S. L., Suleman, L., Vuotto, C. & Donelli, G. Healthcare-associated infections, medical devices and biofilms: risk, tolerance and control. J. Med. Microbiol. 64, 323–334 (2015).
Pulido, L., Ghanem, E., Joshi, A., Purtill, J. J. & Parvizi, J. Periprosthetic joint infection: the incidence, timing, and predisposing factors. Clin. Orthop. Relat. Res. 466, 1710–1715 (2008).
Arciola, C. R., Campoccia, D. & Montanaro, L. Implant infections: adhesion, biofilm formation and immune evasion. Nat. Rev. Microbiol. 16, 397–409 (2018).
Flemming, H. C. et al. Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 14, 563–575 (2016).
Balaban, N. Q. et al. Definitions and guidelines for research on antibiotic persistence. Nat. Rev. Microbiol. 17, 441–448 (2019).
Cameron, D. R., Shan, Y., Zalis, E. A., Isabella, V. & Lewis, K. A genetic determinant of persister cell formation in bacterial pathogens. J. Bacteriol. 200, e00303-18 (2018).
Scherr, T. D. et al. Staphylococcus aureus biofilms induce macrophage dysfunction through leukocidin AB and alpha-toxin. mBio 6, e01021-15 (2015).
Koziel, J. et al. The Janus face of ɑ-toxin: a potent mediator of cytoprotection in staphylococci-infected macrophages. J. Innate Immun. 7, 187–198 (2015).
Schommer, N. N. et al. Staphylococcus epidermidis uses distinct mechanisms of biofilm formation to interfere with phagocytosis and activation of mouse macrophage-like cells 774A.1. Infect. Immun. 79, 2267–2276 (2011).
Thurlow, L. R. et al. Staphylococcus aureus biofilms prevent macrophage phagocytosis and attenuate inflammation in vivo. J. Immunol. 186, 6585–6596 (2011).
Ricciardi, B. F. et al. Staphylococcus aureus evasion of host immunity in the setting of prosthetic joint infection: biofilm and beyond. Curr. Rev. Musculoskelet. Med. 11, 389–400 (2018).
Le, K. Y., Park, M. D. & Otto, M. Immune evasion mechanisms of Staphylococcus epidermidis biofilm infection. Front. Microbiol. 9, 359 (2018).
He, L. et al. Resistance to leukocytes ties benefits of quorum sensing dysfunctionality to biofilm infection. Nat. Microbiol. 4, 1114–1119 (2019).
Heim, C. E. et al. Myeloid-derived suppressor cells contribute to Staphylococcus aureus orthopedic biofilm infection. J. Immunol. 192, 3778–3792 (2014).
Heim, C. E. et al. IL-12 promotes myeloid-derived suppressor cell recruitment and bacterial persistence during Staphylococcus aureus orthopedic implant infection. J. Immunol. 194, 3861–3872 (2015).
Heim, C. E., Vidlak, D. & Kielian, T. Interleukin-10 production by myeloid-derived suppressor cells contributes to bacterial persistence during Staphylococcus aureus orthopedic biofilm infection. J. Leukoc. Biol. 98, 1003–1013 (2015).
Tebartz, C. et al. A major role for myeloid-derived suppressor cells and a minor role for regulatory T cells in immunosuppression during Staphylococcus aureus infection. J. Immunol. 194, 1100–1111 (2015).
Bernthal, N. M. et al. Protective role of IL-1β against post-arthroplasty Staphylococcus aureus infection. J. Orthop. Res. 29, 1621–1626 (2011).
Ouyang, W. & O’Garra, A. IL-10 family cytokines IL-10 and IL-22: from basic science to clinical translation. Immunity 50, 871–891 (2019).
Kessler, B. et al. Interleukin 10 inhibits pro-inflammatory cytokine responses and killing of Burkholderia pseudomallei. Sci. Rep. 7, 42791 (2017).
Leech, J. M., Lacey, K. A., Mulcahy, M. E., Medina, E. & McLoughlin, R. M. IL-10 plays opposing roles during Staphylococcus aureus systemic and localized infections. J. Immunol. 198, 2352–2365 (2017).
Alter, G. et al. IL-10 induces aberrant deletion of dendritic cells by natural killer cells in the context of HIV infection. J. Clin. Invest. 120, 1905–1913 (2010).
Smith, L. K. et al. Interleukin-10 directly inhibits CD8+ T cell function by enhancing N-glycan branching to decrease antigen sensitivity. Immunity 48, 299–312 (2018).
Akdis, C. A., Joss, A., Akdis, M., Faith, A. & Blaser, K. A molecular basis for T cell suppression by IL-10: CD28-associated IL-10 receptor inhibits CD28 tyrosine phosphorylation and phosphatidylinositol 3-kinase binding. FASEB J. 14, 1666–1668 (2000).
Liu, B., Tonkonogy, S. L. & Sartor, R. B. Antigen-presenting cell production of IL-10 inhibits T-helper 1 and 17 cell responses and suppresses colitis in mice. Gastroenterology 141, 653–662 (2011).
Sinha, P., Clements, V. K., Bunt, S. K., Albelda, S. M. & Ostrand-Rosenberg, S. Cross-talk between myeloid-derived suppressor cells and macrophages subverts tumor immunity toward a type 2 response. J. Immunol. 179, 977–983 (2007).
Beury, D. W. et al. Cross-talk among myeloid-derived suppressor cells, macrophages, and tumor cells impacts the inflammatory milieu of solid tumors. J. Leukoc. Biol. 96, 1109–1118 (2014).
Fey, P. D. et al. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 4, e00537-12 (2013).
Fuller, J. R. et al. Identification of a lactate-quinone oxidoreductase in Staphylococcus aureus that is essential for virulence. Front. Cell. Infect. Microbiol. 1, 19 (2011).
Stockland, A. E. & San Clemente, C. L. Multiple forms of lactate dehydrogenase in Staphylococcus aureus. J. Bacteriol. 100, 347–353 (1969).
Kondoh, Y., Kawase, M., Kawakami, Y. & Ohmori, S. Concentrations of d-lactate and its related metabolic intermediates in liver, blood, and muscle of diabetic and starved rats. Res. Exp. Med. (Berl.) 192, 407–414 (1992).
Puig-Kroger, A. et al. Peritoneal dialysis solutions inhibit the differentiation and maturation of human monocyte-derived dendritic cells: effect of lactate and glucose-degradation products. J. Leukoc. Biol. 73, 482–492 (2003).
Husain, Z., Huang, Y., Seth, P. & Sukhatme, V. P. Tumor-derived lactate modifies antitumor immune response: effect on myeloid-derived suppressor cells and NK cells. J. Immunol. 191, 1486–1495 (2013).
Zhang, D. et al. Metabolic regulation of gene expression by histone lactylation. Nature 574, 575–580 (2019).
Ratter, J. M. et al. In vitro and in vivo effects of lactate on metabolism and cytokine production of human primary PBMCs and monocytes. Front. Immunol. 9, 2564 (2018).
Latham, T. et al. Lactate, a product of glycolytic metabolism, inhibits histone deacetylase activity and promotes changes in gene expression. Nucleic Acids Res. 40, 4794–4803 (2012).
Wagner, W., Ciszewski, W. M. & Kania, K. D. l- and d-lactate enhance DNA repair and modulate the resistance of cervical carcinoma cells to anticancer drugs via histone deacetylase inhibition and hydroxycarboxylic acid receptor 1 activation. Cell Commun. Signal. 13, 36 (2015).
Strahl, B. D. & Allis, C. D. The language of covalent histone modifications. Nature 403, 41–45 (2000).
Shakespear, M. R., Halili, M. A., Irvine, K. M., Fairlie, D. P. & Sweet, M. J. Histone deacetylases as regulators of inflammation and immunity. Trends Immunol. 32, 335–343 (2011).
Cheng, F. et al. Divergent roles of histone deacetylase 6 (HDAC6) and histone deacetylase 11 (HDAC11) on the transcriptional regulation of IL10 in antigen presenting cells. Mol. Immunol. 60, 44–53 (2014).
Villagra, A. et al. The histone deacetylase HDAC11 regulates the expression of interleukin 10 and immune tolerance. Nat. Immunol. 10, 92–100 (2009).
Garvie, E. I. Bacterial lactate dehydrogenases. Microbiol. Rev. 44, 106–139 (1980).
Gaspar, P., Al-Bayati, F. A., Andrew, P. W., Neves, A. R. & Yesilkaya, H. Lactate dehydrogenase is the key enzyme for pneumococcal pyruvate metabolism and pneumococcal survival in blood. Infect. Immun. 82, 5099–5109 (2014).
Bunch, P. K., Mat-Jan, F., Lee, N. & Clark, D. P. The ldhA gene encoding the fermentative lactate dehydrogenase of Escherichia coli. Microbiology 143, 187–195 (1997).
Feldman-Salit, A. et al. Regulation of the activity of lactate dehydrogenases from four lactic acid bacteria. J. Biol. Chem. 288, 21295–21306 (2013).
Spahich, N. A., Vitko, N. P., Thurlow, L. R., Temple, B. & Richardson, A. R. Staphylococcus aureus lactate- and malate-quinone oxidoreductases contribute to nitric oxide resistance and virulence. Mol. Microbiol. 100, 759–773 (2016).
Bola, B. M. et al. Inhibition of monocarboxylate transporter-1 (MCT1) by AZD3965 enhances radiosensitivity by reducing lactate transport. Mol. Cancer Ther. 13, 2805–2816 (2014).
Wang, Q. et al. Characterization of monocarboxylate transport in human kidney HK-2 cells. Mol. Pharm. 3, 675–685 (2006).
Polanski, R. et al. Activity of the monocarboxylate transporter 1 inhibitor AZD3965 in small cell lung cancer. Clin. Cancer Res. 20, 926–937 (2014).
Corbet, C. et al. Interruption of lactate uptake by inhibiting mitochondrial pyruvate transport unravels direct antitumor and radiosensitizing effects. Nat. Commun. 9, 1208 (2018).
Garcia-Castillo, V. et al. Targeting metabolic remodeling in triple negative breast cancer in a murine model. J. Cancer 8, 178–189 (2017).
Zhao, Z., Han, F., Yang, S., Wu, J. & Zhan, W. Oxamate-mediated inhibition of lactate dehydrogenase induces protective autophagy in gastric cancer cells: involvement of the Akt-mTOR signaling pathway. Cancer Lett. 358, 17–26 (2015).
Zhai, X., Yang, Y., Wan, J., Zhu, R. & Wu, Y. Inhibition of LDH-A by oxamate induces G2/M arrest, apoptosis and increases radiosensitivity in nasopharyngeal carcinoma cells. Oncol. Rep. 30, 2983–2991 (2013).
Askarian, F., Wagner, T., Johannessen, M. & Nizet, V. Staphylococcus aureus modulation of innate immune responses through Toll-like (TLR), (NOD)-like (NLR) and C-type lectin (CLR) receptors. FEMS Microbiol. Rev. 42, 656–671 (2018).
Hanzelmann, D. et al. Toll-like receptor 2 activation depends on lipopeptide shedding by bacterial surfactants. Nat. Commun. 7, 12304 (2016).
Kawasaki, T. & Kawai, T. Toll-like receptor signaling pathways. Front. Immunol. 5, 461 (2014).
Larsson, L., Thorbert-Mros, S., Rymo, L. & Berglundh, T. Influence of epigenetic modifications of the interleukin-10 promoter on IL10 gene expression. Eur. J. Oral Sci. 120, 14–20 (2012).
Yuan, Z. L., Guan, Y. J., Chatterjee, D. & Chin, Y. E. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science 307, 269–273 (2005).
Qin, G. et al. Metformin blocks myeloid-derived suppressor cell accumulation through AMPK-DACH1-CXCL1 axis. Oncoimmunology 7, e1442167 (2018).
Ouzounova, M. et al. Monocytic and granulocytic myeloid derived suppressor cells differentially regulate spatiotemporal tumour plasticity during metastatic cascade. Nat. Commun. 8, 14979 (2017).
Zhang, C. X. et al. STING signaling remodels the tumor microenvironment by antagonizing myeloid-derived suppressor cell expansion. Cell Death Differ. 26, 2314–2328 (2019).
Sundaram, K. et al. IκBζ regulates human monocyte pro-inflammatory responses induced by Streptococcus pneumoniae. PLoS ONE 11, e0161931 (2016).
Katoh, H. et al. CXCR2-expressing myeloid-derived suppressor cells are essential to promote colitis-associated tumorigenesis. Cancer Cell 24, 631–644 (2013).
Li, W., Mao, Z., Fan, X., Cui, L. & Wang, X. Cyclooxygenase 2 promoted the tumorigenecity of pancreatic cancer cells. Tumour Biol. 35, 2271–2278 (2014).
Lucas, M., Zhang, X., Prasanna, V. & Mosser, D. M. ERK activation following macrophage FcγR ligation leads to chromatin modifications at the IL-10 locus. J. Immunol. 175, 469–477 (2005).
Turner, B. M. Cellular memory and the histone code. Cell 111, 285–291 (2002).
Youn, J. I. et al. Epigenetic silencing of retinoblastoma gene regulates pathologic differentiation of myeloid cells in cancer. Nat. Immunol. 14, 211–220 (2013).
Rosborough, B. R., Castellaneta, A., Natarajan, S., Thomson, A. W. & Turnquist, H. R. Histone deacetylase inhibition facilitates GM-CSF-mediated expansion of myeloid-derived suppressor cells in vitro and in vivo. J. Leukoc. Biol. 91, 701–709 (2012).
Sahakian, E. et al. Histone deacetylase 11: a novel epigenetic regulator of myeloid derived suppressor cell expansion and function. Mol. Immunol. 63, 579–585 (2015).
Eskandarian, H. A. et al. A role for SIRT2-dependent histone H3K18 deacetylation in bacterial infection. Science 341, 1238858 (2013).
Kincaid, E. Z. & Ernst, J. D. Mycobacterium tuberculosis exerts gene-selective inhibition of transcriptional responses to IFN-γ without inhibiting STAT1 function. J. Immunol. 171, 2042–2049 (2003).
Wang, Y., Curry, H. M., Zwilling, B. S. & Lafuse, W. P. Mycobacteria inhibition of IFN-γ induced HLA-DR gene expression by up-regulating histone deacetylation at the promoter region in human THP-1 monocytic cells. J. Immunol. 174, 5687–5694 (2005).
Pathak, S. K. et al. TLR4-dependent NF-κB activation and mitogen- and stress-activated protein kinase 1-triggered phosphorylation events are central to Helicobacter pylori peptidyl prolyl cis-, trans-isomerase (HP0175)-mediated induction of IL-6 release from macrophages. J. Immunol. 177, 7950–7958 (2006).
Cao, J. et al. HDAC11 regulates type I interferon signaling through defatty-acylation of SHMT2. Proc. Natl Acad. Sci. USA 116, 5487–5492 (2019).
Moreno-Yruela, C., Galleano, I., Madsen, A. S. & Olsen, C. A. Histone deacetylase 11 is an ε-N-myristoyllysine hydrolase. Cell Chem. Biol. 25, 849–856 e848 (2018).
Kutil, Z. et al. Histone deacetylase 11 is a fatty-acid deacylase. ACS Chem. Biol. 13, 685–693 (2018).
Korkmaz, B., Horwitz, M. S., Jenne, D. E. & Gauthier, F. Neutrophil elastase, proteinase 3, and cathepsin G as therapeutic targets in human diseases. Pharm. Rev. 62, 726–759 (2010).
Lau, D. et al. Myeloperoxidase mediates neutrophil activation by association with CD11b/CD18 integrins. Proc. Natl Acad. Sci. USA 102, 431–436 (2005).
Kumar, V., Patel, S., Tcyganov, E. & Gabrilovich, D. I. The nature of myeloid-derived suppressor cells in the tumor microenvironment. Trends Immunol. 37, 208–220 (2016).
Obermajer, N. & Kalinski, P. Key role of the positive feedback between PGE(2) and COX2 in the biology of myeloid-derived suppressor cells. Oncoimmunology 1, 762–764 (2012).
Dufait, I. et al. Perforin and granzyme B expressed by murine myeloid-derived suppressor cells: a study on their role in outgrowth of cancer cells. Cancers (Basel) 11, 808 (2019).
Sawant, A. et al. Myeloid-derived suppressor cells function as novel osteoclast progenitors enhancing bone loss in breast cancer. Cancer Res. 73, 672–682 (2013).
Heim, C. E. et al. Human prosthetic joint infections are associated with myeloid-derived suppressor cells (MDSCs): implications for infection persistence. J. Orthop. Res. 36, 1605–1613 (2018).
Manning Fox, J. E., Meredith, D. & Halestrap, A. P. Characterisation of human monocarboxylate transporter 4 substantiates its role in lactic acid efflux from skeletal muscle. J. Physiol. 529, 285–293 (2000).
Vitko, N. P., Grosser, M. R., Khatri, D., Lance, T. R. & Richardson, A. R. Expanded glucose import capability affords Staphylococcus aureus optimized glycolytic flux during infection. mBio 7, e00296-16 (2016).
Vitko, N. P., Spahich, N. A. & Richardson, A. R. Glycolytic dependency of high-level nitric oxide resistance and virulence in Staphylococcus aureus. mBio 6, e00045-15 (2015).
Yermak, K., Karbysheva, S., Perka, C., Trampuz, A. & Renz, N. Performance of synovial fluid d-lactate for the diagnosis of periprosthetic joint infection: a prospective observational study. J. Infect. 79, 123–129 (2019).
Zhang, Q. & Cao, X. Epigenetic regulation of the innate immune response to infection. Nat. Rev. Immunol. 19, 417–432 (2019).
Turner, N. A. et al. Methicillin-resistant Staphylococcus aureus: an overview of basic and clinical research. Nat. Rev. Microbiol. 17, 203–218 (2019).
Sun, L. et al. Loss of HDAC11 ameliorates clinical symptoms in a multiple sclerosis mouse model. Life Sci. Alliance 1, e201800039 (2018).
Mootz, J. M., Malone, C. L., Shaw, L. N. & Horswill, A. R. Staphopains modulate Staphylococcus aureus biofilm integrity. Infect. Immun. 81, 3227–3238 (2013).
Gries, C. M. et al. Cyclic di-AMP released from Staphylococcus aureus biofilm induces a macrophage type I interferon response. Infect. Immun. 84, 3564–3574 (2016).
Yamada, K. J. et al. Arginase-1 expression in myeloid cells regulates Staphylococcus aureus planktonic but not biofilm infection. Infect. Immun. 86, e00206-18 (2018).
Heim, C. E., West, S. C., Ali, H. & Kielian, T. Heterogeneity of Ly6G+ Ly6C+ myeloid-derived suppressor cell infiltrates during Staphylococcus aureus biofilm infection. Infect. Immun. 86, e00684-18 (2018).
Niska, J. A. et al. Vancomycin-rifampin combination therapy has enhanced efficacy against an experimental Staphylococcus aureus prosthetic joint infection. Antimicrob. Agents Chemother. 57, 5080–5086 (2013).
Pribaz, J. R. et al. Mouse model of chronic post-arthroplasty infection: noninvasive in vivo bioluminescence imaging to monitor bacterial burden for long-term study. J. Orthop. Res. 30, 335–340 (2012).
Niska, J. A. et al. Monitoring bacterial burden, inflammation and bone damage longitudinally using optical and muCT imaging in an orthopaedic implant infection in mice. PLoS ONE 7, e47397 (2012).
Yamada, K. J. et al. Monocyte metabolic reprogramming promotes pro-inflammatory activity and Staphylococcus aureus biofilm clearance. PLoS Pathog. 16, e1008354 (2020).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Picard Toolkit (Broad Institute, GitHub repository, 2019); http://broadinstitute.github.io/picard/
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Allhoff, M., Sere, K., J, F. P., Zenke, M. & I, G. C. Differential peak calling of ChIP-seq signals with replicates with THOR. Nucleic Acids Res. 44, e153 (2016).
Allhoff, M. et al. Detecting differential peaks in ChIP-seq signals with ODIN. Bioinformatics 30, 3467–3475 (2014).
Ramírez, F. et al. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–W191 (2014).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Lin, X., Tirichine, L. & Bowler, C. Protocol: chromatin immunoprecipitation (ChIP) methodology to investigate histone modifications in two model diatom species. Plant Methods 8, 48 (2012).
Pfaffl, M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45 (2001).
Trim Galore (Babraham Institute, GitHub repository, 2019); https://github.com/FelixKrueger/TrimGalore
Trapnell, C., Pachter, L. & Salzberg, S. L. TopHat: discovering splice junctions with RNA-seq. Bioinformatics 25, 1105–1111 (2009).
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protocols 7, 562–578 (2012).
Acknowledgements
This work was supported by the National Institutes of Health/National Institute of Allergy and Infectious Diseases grant no. P01AI083211 (Project 4 to T.K.) and grant no. R01AI125588 (to V.C.T.). The authors thank R. Fallet for managing the mouse colony. The UNMC DNA Sequencing Core receives partial support from the National Institute for General Medical Science (grant nos. INBRE–P20GM103427-14 and COBRE–1P30GM110768-01). Both the UNMC DNA Sequencing and Flow Cytometry Research Cores receive support from The Fred & Pamela Buffett Cancer Center Support Grant (grant no. P30CA036727).
Author information
Authors and Affiliations
Contributions
C.E.H. and T.K. conceived the study; C.E.H., M.E.B., K.J.Y., A.L.A. and T.K. designed experiments; and A.R.K. and D.K. provided expertise in the design, execution and data analysis for the ChIP–seq, ChIP–PCR and scRNA-seq experiments. S.S.C., A.A.A., C.M.G. and V.C.T. created the S. aureus lactate mutants used in the study. C.E.H., M.E.B., K.J.Y. and A.L.A. conducted the in vivo mouse PJI experiments. C.E.H. performed the in vitro biofilm–leukocyte co-culture experiments. E.S. and Y.L. provided the HDAC11 KO mice. C.E.H., D.K. and T.K. performed data analysis. T.K. procured funding for this work. C.E.H. and T.K. wrote the manuscript. All authors edited and approved the submission of this work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 S. aureus lactate mutants do not display growth defects in liquid broth or biofilm in vitro.
a, S. aureus strains used in this study. b, The growth rate of S. aureus WT, Δddh, Δldh1/ldh2, and Δddh/ldh1/ldh2 was determined in brain-heart infusion broth over a 24 h period with constant agitation using a TECAN (7 biological replicates/strain). c, Strains were transduced with a sarA-GFP plasmid and grown for 4 days under static growth conditions in RPMI-1640 supplemented with 10% FBS, whereupon biofilm formation was visualized by confocal microscopy. Results are representative of two independent experiments, each with 4 biological replicates. Scale bars, 100 µm.
Extended Data Fig. 2 Intracellular pH of MDSCs and macrophages is not dramatically altered by S. aureus-derived lactate during biofilm co-culture.
MDSCs or macrophages were labeled with BCECF-AM (10 µM) prior to co-culture with WT (n = 4 biological replicates/group) or Δddh/ldh1/ldh2 (n = 4 and 3 biological replicates for MDSCs and macrophages, respectively) biofilm for 2 h, whereupon intracellular pH was determined by flow cytometry based on a standard curve of known pH. Results shown are from one experiment.
Extended Data Fig. 3 D- and L-lactate production during S. aureus orthopaedic infection.
(a) L- and (b) D-lactate were quantified in the implant-associated tissue of mice infected with WT, Δddh, Δldh1/ldh2, or Δddh/ldh1/ldh2 at days 3, 14, and 28 post-infection (mean ± SD; n = 5/group). The dashed line represents background in the assay as determined with tissues collected from animals receiving sterile implants at the same time points (n = 5 at days 3 and 28 and n = 4 at day 14). Results are representative of three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001; One-way ANOVA.
Extended Data Fig. 4 The expression of select inflammatory mediators is independent of bacterial burden during S. aureus orthopaedic infection.
Cytokine levels were quantified in implant-associated tissue of mice infected with WT, Δddh/ldh1/ldh2, or a 1:1 ratio of WT and Δddh/ldh1/ldh2 at days 3, 14, and 28 post-infection. Results are combined from two independent experiments (mean ± SD; n = 8/group). *, p < 0.05; One-way ANOVA.
Extended Data Fig. 5 IL-10 production during S. aureus orthopaedic infection is not influenced by host lactate.
Mice received daily i.p. injections of sodium oxamate (500 mg/kg/day) dissolved in 0.5% Hydroxypropyl Methylcellulose or vehicle (0.5% Hydroxypropyl Methylcellulose) beginning one day prior to infection with S. aureus WT (n = 10/group) or Δddh/ldh1/ldh2 (n = 10 or 9 for vehicle and oxamate, respectively). Mice were killed at day 14 post-infection to quantify (a) D-lactate, (b) L-lactate, and (c) IL-10 in implant-associated tissue. Results are combined from two independent experiments (mean ± SD). D- and L-lactate measurements are also reported at day 14 for mice that received sterile implants (n = 4/group). *, p < 0.05; **, p < 0.01; ***, p < 0.001; One-way ANOVA. NS, not significant.
Extended Data Fig. 6 Sodium oxamate does not affect S. aureus growth or lactate production.
a, D- and (b) L-lactate were quantified in S. aureus WT and Δddh/ldh1/ldh2 biofilm in 96-well plates under static growth conditions in RPMI-1640 supplemented with 10% FBS over a 4 d period. The dotted lines represent daily medium changes. Results are from one experiment with 5 biological replicates. S. aureus was exposed to various concentrations of sodium oxamate during (c) growth in liquid broth (brain-heart infusion) beginning at time 0 (n = 6 biological replicates/group) or (d) throughout the 4-day biofilm maturation period (n = 5 biological replicates/group). Biofilm cultures were replenished daily with fresh medium (RPMI-1640 + 10% FBS) containing sodium oxamate. Results are presented as (c) OD600 or (d) Log10 colony forming units (CFU) per well. e, Quantification of D- and L-lactate from biofilm throughout the 4-day growth period, where the dotted lines represent daily medium changes (n = 4 biological replicates/group). All results are reported as mean ± SD.
Extended Data Fig. 7 Effects of S. aureus lactate on orthopaedic implant biofilm infection are IL-10-dependent.
WT and IL-10 KO mice were infected with WT (n = 10) or Δddh/ldh1/ldh2 (n = 9) S. aureus, whereupon bacterial burden in (a) implant-associated tissue and (b) femur as well as (c) MDSC and (d) monocyte infiltrates were assessed at day 14 post-infection. Results represent the mean ± SEM of two independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001; One-way ANOVA.
Extended Data Fig. 8 S. aureus-derived lactate inhibits the negative regulator HDAC11 to augment leukocyte IL-10 production in a HDAC6-dependent manner.
MDSCs and macrophages from WT or HDAC11 KO mice were co-cultured with (a) WT or (b) Δddh/ldh1/ldh2 biofilm for 2 h ± HDAC6i (36 nM). IL-10 production was measured by cytometric bead array. Results represent the mean ± SEM of two independent experiments (n = 8 and 12 biological replicates for MDSCs and macrophages, respectively). **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; One-way ANOVA. Values for untreated leukocytes are the same as those presented in Fig. 5a,b because both HDAC6i and tubastatin A were tested at the same time.
Extended Data Fig. 9 S. aureus-derived lactate preferentially inhibits HDAC11.
Purified active HDAC11 or HDAC6 were exposed to conditioned medium from WT or Δddh/ldh1/ldh2 biofilm for 30 min, whereupon HDAC activity was determined using a fluorescent HDAC substrate (deAc-FdL). Results are from one experiment with 4 biological replicates and are expressed as the percent change in HDAC activity compared to purified enzyme. **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; One-way ANOVA. NS, not significant.
Extended Data Fig. 10 Gating strategy to quantitate leukocyte populations in S. aureus implant-associated soft tissue.
Single cells were gated from the (a) total events using (b) FSC-A vs. FSC-H, followed by (c) exclusion of dead cells. (d) Live, CD45+ leukocytes were separated into (e) Ly6G+Ly6C+ vs. Ly6G−Ly6C+. (f) MDSC and neutrophil populations were identified based on CD11b expression, while (g) monocyte and macrophage populations were identified based on Ly6C and F4/80 expression, respectively.
Supplementary information
Supplementary Information
Supplementary Figs. 1–4 and Tables 1 and 2.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Rights and permissions
About this article
Cite this article
Heim, C.E., Bosch, M.E., Yamada, K.J. et al. Lactate production by Staphylococcus aureus biofilm inhibits HDAC11 to reprogramme the host immune response during persistent infection. Nat Microbiol 5, 1271–1284 (2020). https://doi.org/10.1038/s41564-020-0756-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-020-0756-3
This article is cited by
-
Transcriptional Expression of Histone Acetyltransferases and Deacetylases During the Recovery of Acute Exercise in Mouse Hippocampus
Journal of Molecular Neuroscience (2024)
-
Single-cell transcriptome reveals Staphylococcus aureus modulating fibroblast differentiation in the bone-implant interface
Molecular Medicine (2023)
-
IL-10 production by granulocytes promotes Staphylococcus aureus craniotomy infection
Journal of Neuroinflammation (2023)
-
Staphylococcus aureus host interactions and adaptation
Nature Reviews Microbiology (2023)
-
Bacterial and Metabolic Factors of Staphylococcal Planktonic and Biofilm Environments Differentially Regulate Macrophage Immune Activation
Inflammation (2023)