Abstract
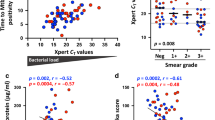
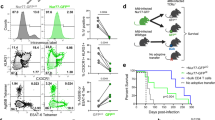
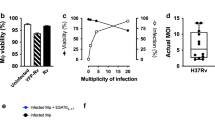
Mycobacterium tuberculosis infection (Mtb) is the leading cause of death due to a single infectious agent and is among the top ten causes of all human deaths worldwide1. CD4 T cells are essential for resistance to Mtb infection, and for decades it has been thought that IFNγ production is the primary mechanism of CD4 T-cell-mediated protection2,3. However, IFNγ responses do not correlate with host protection, and several reports demonstrate that additional anti-tuberculosis CD4 T-cell effector functions remain unaccounted for4,5,6,7,8. Here we show that the tumour-necrosis factor (TNF) superfamily molecule CD153 (encoded by the gene Tnfsf8) is required for control of pulmonary Mtb infection by CD4 T cells. In Mtb-infected mice, CD153 expression is highest on Mtb-specific T helper 1 (TH1) cells in the lung tissue parenchyma, but its induction does not require TH1 cell polarization. CD153-deficient mice develop high pulmonary bacterial loads and succumb early to Mtb infection. Reconstitution of T-cell-deficient hosts with either Tnfsf8−/− or Ifng−/− CD4 T cells alone fails to rescue mice from early mortality, but reconstitution with a mixture of Tnfsf8−/− and Ifng−/− CD4 T cells provides similar protection as wild-type T cells. In Mtb-infected non-human primates, CD153 expression is much higher on Ag-specific CD4 T cells in the airways compared to blood, and the frequency of Mtb-specific CD153-expressing CD4 T cells inversely correlates with bacterial loads in granulomas. In Mtb-infected humans, CD153 defines a subset of highly polyfunctional Mtb-specific CD4 T cells that are much more abundant in individuals with controlled latent Mtb infection compared to those with active tuberculosis. In all three species, Mtb-specific CD8 T cells did not upregulate CD153 following peptide stimulation. Thus, CD153 is a major immune mediator of host protection against pulmonary Mtb infection and CD4 T cells are one important source of this molecule.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
CD4 T-cell microarray data are publicly available, accession number GSE116830. All data reported in this study are available from the corresponding author upon reasonable request.
References
WHO. Global Tuberculosis Report 2016. http://www.who.int/tb/publications/global_report/gtbr2016_executive_summary.pdf (World Health Organization, Geneva, 2016).
Cooper, A. M. et al. Disseminated tuberculosis in interferon gamma gene-disrupted mice. J. Exp. Med. 178, 2243–2247 (1993).
Flynn, J. L. et al. An essential role for interferon gamma in resistance to Mycobacterium tuberculosis infection. J. Exp. Med. 178, 2249–2254 (1993).
Cowley, S. C. & Elkins, K. L. CD4+ T cells mediate IFN-γ-independent control of Mycobacterium tuberculosis infection both in vitro and in vivo. J. Immunol. 171, 4689–4699 (2003).
Gallegos, A. M. et al. A gamma interferon independent mechanism of CD4 T cell mediated control of M. tuberculosis infection in vivo. PLoS Pathog. 7, e1002052 (2011).
Orr, M. T. et al. Mucosal delivery switches the response to an adjuvanted tuberculosis vaccine from systemic TH1 to tissue-resident TH17 responses without impacting the protective efficacy. Vaccine 33, 6570–6578 (2015).
Orr, M. T. et al. Interferon γ and tumor necrosis factor are not essential parameters of CD4+ T-cell responses for vaccine control of tuberculosis. J. Infect. Dis. 212, 495–504 (2015).
Sakai, S. et al. CD4 T cell-derived IFN-γ plays a minimal role in control of pulmonary Mycobacterium tuberculosis infection and must be actively repressed by PD-1 to prevent lethal disease. PLoS Pathog. 12, e1005667 (2016).
Sallin, M. A. et al. TH1 differentiation drives the accumulation of intravascular, non-protective CD4 T cells during tuberculosis. Cell Rep. 18, 3091–3104 (2017).
Moguche, A. O. et al. ICOS and Bcl6-dependent pathways maintain a CD4 T cell population with memory-like properties during tuberculosis. J. Exp. Med. 212, 715–728 (2015).
Sakai, S. et al. Cutting edge: control of Mycobacterium tuberculosis infection by a subset of lung parenchyma-homing CD4 T cells. J. Immunol. 192, 2965–2969 (2014).
Lazarevic, V., Myers, A. J., Scanga, C. A. & Flynn, J. L. CD40, but not CD40L, is required for the optimal priming of T cells and control of aerosol M. tuberculosis infection. Immunity 19, 823–835 (2003).
Musicki, K., Briscoe, H., Britton, W. J. & Saunders, B. M. LIGHT contributes to early but not late control of Mycobacterium tuberculosis infection. Int. Immunol. 22, 353–358 (2010).
Allie, N. et al. Limited role for lymphotoxin α in the host immune response to Mycobacterium tuberculosis. J. Immunol. 185, 4292–4301 (2010).
Martinez Gomez, J. M. et al. Role of the CD137 ligand (CD137L) signaling pathway during Mycobacterium tuberculosis infection. Immunobiology 219, 78–86 (2014).
Tang, C. et al. A novel role of CD30L/CD30 signaling by T–T cell interaction in TH1 response against mycobacterial infection. J. Immunol. 181, 6316–6327 (2008).
Marin, N. D. & Garcia, L. F. The role of CD30 and CD153 (CD30L) in the anti-mycobacterial immune response. Tuberculosis 102, 8–15 (2017).
Florido, M., McColl, S. R. & Appelberg, R. Delayed recruitment of lymphocytes into the lungs of CD30-deficient mice during aerogenic Mycobacterium avium infections. Immunobiology 214, 643–652 (2009).
Florido, M., Borges, M., Yagita, H. & Appelberg, R. Contribution of CD30/CD153 but not of CD27/CD70, CD134/OX40L, or CD137/4-1BBL to the optimal induction of protective immunity to Mycobacterium avium. J. Leukoc. Biol. 76, 1039–1046 (2004).
Umeda, K. et al. Innate memory phenotype CD4+ T cells play a role in early protection against infection by Listeria monocytogenes in a CD30L-dependent manner. Microbiol. Immunol. 55, 645–656 (2011).
Fava, V. M. et al. Association of TNFSF8 regulatory variants with excessive inflammatory responses but not leprosy per se. J. Infect. Dis. 211, 968–977 (2015).
Torrado, E. et al. Interleukin 27R regulates CD4+ T cell phenotype and impacts protective immunity during Mycobacterium tuberculosis infection. J. Exp. Med. 212, 1449–1463 (2015).
Mothe, B. R. et al. The TB-specific CD4+ T cell immune repertoire in both cynomolgus and rhesus macaques largely overlap with humans. Tuberculosis 95, 722–735 (2015).
Lindestam Arlehamn, C. S. et al. A quantitative analysis of complexity of human pathogen-specific CD4 T cell responses in healthy M. tuberculosis infected South Africans. PLoS Pathog. 12, e1005760 (2016).
Strickland, N. et al. Characterization of Mycobacterium tuberculosis-specific cells using MHC class II tetramers reveals phenotypic differences related to HIV infection and tuberculosis disease. J. Immunol. 199, 2440–2450 (2017).
Riou, C., Berkowitz, N., Goliath, R., Burgers, W. A. & Wilkinson, R. J. Analysis of the phenotype of Mycobacterium tuberculosis-specific CD4+ T cells to discriminate latent from active tuberculosis in HIV-uninfected and HIV-infected individuals. Front. Immunol. 8, 968 (2017).
Adekambi, T. et al. Biomarkers on patient T cells diagnose active tuberculosis and monitor treatment response. J. Clin. Invest. 125, 1827–1838 (2015).
Day, C. L. et al. Functional capacity of Mycobacterium tuberculosis-specific T cell responses in humans is associated with mycobacterial load. J. Immunol. 187, 2222–2232 (2011).
Harari, A. et al. Dominant TNF-α+ Mycobacterium tuberculosis-specific CD4+ T cell responses discriminate between latent infection and active disease. Nat. Med. 17, 372–376 (2011).
Pollock, K. M. et al. T-cell immunophenotyping distinguishes active from latent tuberculosis. J. Infect. Dis. 208, 952–968 (2013).
Wilkinson, K. A. et al. Activation profile of Mycobacterium tuberculosis-specific CD4+ T cells reflects disease activity irrespective of HIV status. Am. J. Respir. Crit. Care Med. 193, 1307–1310 (2016).
Halliday, A. et al. Stratification of latent Mycobacterium tuberculosis infection by cellular immune profiling. J. Infect. Dis. 215, 1480–1487 (2017).
Lichtner, M. et al. Multifunctional analysis of CD4+ T-cell response as immune-based model for tuberculosis detection. J. Immunol. Res. 2015, 217–287 (2015).
Qiu, Z. et al. Multifunctional CD4 T cell responses in patients with active tuberculosis. Sci. Rep. 2, 216 (2012).
Sutherland, J. S., Adetifa, I. M., Hill, P. C., Adegbola, R. A. & Ota, M. O. Pattern and diversity of cytokine production differentiates between Mycobacterium tuberculosis infection and disease. Eur. J. Immunol. 39, 723–729 (2009).
Harris, R. C., Sumner, T., Knight, G. M. & White, R. G. Systematic review of mathematical models exploring the epidemiological impact of future TB vaccines. Hum. Vaccin. Immunother. 12, 2813–2832 (2016).
Kauffman, K. D. et al. Defective positioning in granulomas but not lung-homing limits CD4 T-cell interactions with Mycobacterium tuberculosis-infected macrophages in rhesus macaques. Mucosal Immunol. 11, 462–473 (2018).
Acknowledgements
We thank N. Zhu for technical assistance and the animal care staff of National Institute of Allergy and Infectious Diseases (NIAID) Comparative Medicine Branch. We thank B. Hague for help with cell sorting. We thank R. Germain for helpful discussions. This work was supported by the Intramural Research Programme of NIAID/National Institutes of Health (NIH) and the NIH (5U01AI115940-04). R.J.W. is supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC00110218), the UK Medical Research Council (FC00110218) and Wellcome (FC00110218). R.J.W. also receives support from Wellcome (104803, 203135), NIH (U19A1111276) and the Medical Research Council of South Africa (SHIP).
Author information
Authors and Affiliations
Contributions
D.L.B. conceived the project. D.L.B., M.A.S. and K.D.K. designed the research. M.A.S., K.D.K., S.G.H. and S.S. performed the mouse model experiments. T.G.M. and P.J.G. performed and analysed the microarray experiments. K.D.K., M.A.S., S.S., S.G.H. and T.W.F. performed the NHP model experiments. R.M., T.W.-K. and I.N.M. performed the bronchoscopic Mtb infections and provided veterinary care for infected NHPs under aBSL3 conditions. D.L.B., M.A.S. and K.D.K. analysed the mouse and NHP model data. A.Se. and C.S.L.A. provided critical reagents. R.J.W. and A.Sh. designed the human subjects research. C.R. and E.D.B. performed human subjects research and analysed the data. D.L.B., M.A.S. and K.D.K. wrote the manuscript with all authors contributing to writing and providing feedback.
Corresponding author
Ethics declarations
Competing interests
D.L.B., M.A.S. and K.D.K. are listed as inventors on a patent application filed by NIAID based on these data. The authors declare no other competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–3, Supplementary Tables 1 and 2
Rights and permissions
About this article
Cite this article
Sallin, M.A., Kauffman, K.D., Riou, C. et al. Host resistance to pulmonary Mycobacterium tuberculosis infection requires CD153 expression. Nat Microbiol 3, 1198–1205 (2018). https://doi.org/10.1038/s41564-018-0231-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-018-0231-6
This article is cited by
-
Targeting aging and age-related diseases with vaccines
Nature Aging (2024)
-
Recently activated CD4 T cells in tuberculosis express OX40 as a target for host-directed immunotherapy
Nature Communications (2023)
-
Immune evasion and provocation by Mycobacterium tuberculosis
Nature Reviews Microbiology (2022)
-
Strategies for targeting senescent cells in human disease
Nature Aging (2021)
-
Mycobacterium tuberculosis-specific CD4 T cells expressing CD153 inversely associate with bacterial load and disease severity in human tuberculosis
Mucosal Immunology (2021)