Abstract
Understanding the temporal succession of ecological communities and the underlying mechanisms in response to climate warming is critical for future climate projections. However, despite its fundamental importance in ecology and evolution, little is known about how the Archaea domain responds to warming. Here we showed that experimental warming of a tallgrass prairie ecosystem significantly altered the community structure of soil archaea and reduced their taxonomic and phylogenetic diversity. In contrast to previous observations in bacteria and fungi, we showed convergent succession of the soil archaeal community between warming and control. Although stochastic processes dominated the archaeal community, their relative importance decreased over time. Furthermore, the warming-induced changes in the archaeal community and soil chemistry had significant impacts on ecosystem functioning. Our results imply that, although the detrimental effects of biodiversity loss on ecosystems could be much severer, the soil archaeal community structure would be more predictable in a warmer world.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The DNA sequences of the archaeal 16S rRNA gene amplicons are available in the National Center for Biotechnology Information (NCBI) Sequence Read Archive under project accession number PRJNA861672. The DNA sequences of the bacterial 16S rRNA gene amplicons are under the project accession number PRJNA331185. Raw shotgun metagenomic sequences are deposited in the European Nucleotide Archive (http://www.ebi.ac.uk/ena) under study no. PRJNA533082. The soil physical and chemical attributes, and plant biomass and richness are downloadable online at http://www.ou.edu/ieg/publications/datasets. Silva 132 Ref NR database is available at https://www.arb-silva.de/documentation/release-132/. The Greengene reference dataset is available from the QIIME GitHub repository https://github.com/biocore/qiime-default-reference/blob/master/qiime_default_reference/gg_13_8_otus/rep_set/97_otus.fasta.gz. Source data are provided with this paper.
Code availability
R scripts for statistical analyses and source data are available on GitHub at https://github.com/yazhang2022/OKwarmingsiteArchaea.
References
Woese, C. R., Kandler, O. & Wheelis, M. L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl Acad. Sci. USA 87, 4576–4579 (1990).
Hedlund, B. P., Zhang, C., Wang, F., Rinke, C. & Martin, W. F. Editorial: ecology, metabolism and evolution of archaea-perspectives from Proceedings of the International Workshop on Geo-Omics of Archaea. Front. Microbiol. 12, 827229 (2021).
DeLong, E. F. Archaea in coastal marine environments. Proc. Natl Acad. Sci. USA 89, 5685–5689 (1992).
Fuhrman, J. A., McCallum, K. & Davis, A. A. Novel major archaebacterial group from marine plankton. Nature 356, 148–149 (1992).
DeLong, E. F. Exploring marine planktonic archaea: then and now. Front. Microbiol. 11, 616086 (2021).
Karimi, B. et al. Biogeography of soil bacteria and archaea across France. Sci. Adv. 4, eaat1808 (2018).
Tahon, G., Geesink, P. & Ettema, T. J. G. Expanding archaeal diversity and phylogeny: past, present, and future. Annu. Rev. Microbiol. 75, 359–381 (2021).
Baker, B. J. et al. Diversity, ecology and evolution of Archaea. Nat. Microbiol. 5, 887–900 (2020).
Needham, D. M. & Fuhrman, J. A. Pronounced daily succession of phytoplankton, archaea and bacteria following a spring bloom. Nat. Microbiol. 1, 16005 (2016).
Shu, W. S. & Huang, L. N. Microbial diversity in extreme environments. Nat. Rev. Microbiol. 20, 219–235 (2021).
Adam, P. S., Borrel, G., Brochier-Armanet, C. & Gribaldo, S. The growing tree of Archaea: new perspectives on their diversity, evolution and ecology. ISME J. 11, 2407–2425 (2017).
Zaremba-Niedzwiedzka, K. et al. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 541, 353–358 (2017).
Imachi, H. et al. Isolation of an archaeon at the prokaryote-eukaryote interface. Nature 577, 519–525 (2020).
Liu, Y. et al. Expanded diversity of Asgard archaea and their relationships with eukaryotes. Nature 593, 553–557 (2021).
Angel, R., Soares, M. I., Ungar, E. D. & Gillor, O. Biogeography of soil archaea and bacteria along a steep precipitation gradient. ISME J. 4, 553–563 (2010).
Auguet, J. C., Barberan, A. & Casamayor, E. O. Global ecological patterns in uncultured Archaea. ISME J. 4, 182–190 (2010).
Bates, S. T. et al. Examining the global distribution of dominant archaeal populations in soil. ISME J. 5, 908–917 (2011).
Bar-On, Y. M., Phillips, R. & Milo, R. The biomass distribution on Earth. Proc. Natl Acad. Sci. USA 115, 6506–6511 (2018).
Offre, P., Spang, A. & Schleper, C. Archaea in biogeochemical cycles. Annu. Rev. Microbiol. 67, 437–457 (2013).
Leininger, S. et al. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442, 806–809 (2006).
Danovaro, R., Rastelli, E., Corinaldesi, C., Tangherlini, M. & Dell’Anno, A. Marine archaea and archaeal viruses under global change. F1000Research 6, 1241 (2017).
Goberna, M., Garcia, C., Insam, H., Hernandez, M. T. & Verdu, M. Burning fire-prone Mediterranean shrublands: immediate changes in soil microbial community structure and ecosystem functions. Microb. Ecol. 64, 242–255 (2012).
Gschwendtner, S. et al. Climate change induces shifts in abundance and activity pattern of bacteria and archaea catalyzing major transformation steps in nitrogen turnover in a soil from a mid-European beech forest. PLoS ONE 9, e114278 (2014).
Hayden, H. L. et al. Changes in the microbial community structure of bacteria, archaea and fungi in response to elevated CO2 and warming in an Australian native grassland soil. Environ. Microbiol. 14, 3081–3096 (2012).
Guo, X. et al. Climate warming leads to divergent succession of grassland microbial communities. Nat. Clim. Change 8, 813–818 (2018).
Guo, X. et al. Climate warming accelerates temporal scaling of grassland soil microbial biodiversity. Nat. Ecol. Evol. 3, 612–619 (2019).
Wu, L. et al. Reduction of microbial diversity in grassland soil is driven by long-term climate warming. Nat. Microbiol. 7, 1054–1062 (2022).
Yuan, M. M. et al. Climate warming enhances microbial network complexity and stability. Nat. Clim. Change 11, 343–348 (2021).
Cavicchioli, R. Archaea–timeline of the third domain. Nat. Rev. Microbiol. 9, 51–61 (2011).
Prach, K. & Walker, L. R. Four opportunities for studies of ecological succession. Trends Ecol. Evol. 26, 119–123 (2011).
Xu, X., Sherry, R. A., Niu, S. L., Li, D. J. & Luo, Y. Q. Net primary productivity and rain-use efficiency as affected by warming, altered precipitation, and clipping in a mixed-grass prairie. Glob. Change Biol. 19, 2753–2764 (2013).
Kerou, M., Alves, R. J. E. & Schleper, C. in Bergey’s Manual of Systematics of Archaea and Bacteria (eds Trujillo, M. E. et al.) https://doi.org/10.1002/9781118960608.obm00124 (John Wiley & Sons, 2018).
Nkamga, V. D. & Drancourt, M. in Bergey’s Manual of Systematics of Archaea and Bacteria (eds Trujillo, M. E. et al.) https://doi.org/10.1002/9781118960608.gbm01365 (John Wiley & Sons, 2016).
Zhou, J. et al. High-throughput metagenomic technologies for complex microbial community analysis: open and closed formats. mBio 6, e02288-14 (2015).
Zhou, J. et al. Random sampling process leads to overestimation of beta-diversity of microbial communities. mBio 4, e00324-13 (2013).
Xue, K. et al. Tundra soil carbon is vulnerable to rapid microbial decomposition under climate warming. Nat. Clim. Change 6, 595–600 (2016).
Pecl, G. T. et al. Biodiversity redistribution under climate change: impacts on ecosystems and human well-being. Science 355, eaai9214 (2017).
Cardinale, B. J. et al. Biodiversity loss and its impact on humanity. Nature 486, 59–67 (2012).
Li, D., Miller, J. E. D. & Harrison, S. Climate drives loss of phylogenetic diversity in a grassland community. Proc. Natl Acad. Sci. USA 116, 19989–19994 (2019).
Fei, S. et al. Divergence of species responses to climate change. Sci. Adv. 3, e1603055 (2017).
Bascompte, J., García, M. B., Ortega, R., Rezende, E. L. & Pironon, S. Mutualistic interactions reshuffle the effects of climate change on plants across the tree of life. Sci. Adv. 5, eaav2539 (2019).
Kerou, M. et al. Proteomics and comparative genomics of Nitrososphaera viennensis reveal the core genome and adaptations of archaeal ammonia oxidizers. Proc. Natl Acad. Sci. USA 113, E7937–E7946 (2016).
Taylor, A. E., Giguere, A. T., Zoebelein, C. M., Myrold, D. D. & Bottomley, P. J. Modeling of soil nitrification responses to temperature reveals thermodynamic differences between ammonia-oxidizing activity of archaea and bacteria. ISME J. 11, 896–908 (2017).
Shi, Z. et al. Functional gene array-based ultrasensitive and quantitative detection of microbial populations in complex communities. mSystems 4, e00296-19 (2019).
Ning, D. L. et al. A quantitative framework reveals ecological drivers of grassland microbial community assembly in response to warming. Nat. Commun. 11, 4717 (2020).
Liu, L. et al. Changes in assembly processes of soil microbial communities during secondary succession in two subtropical forests. Soil Biol. Biochem. 154, 108144 (2021).
Verhamme, D. T., Prosser, J. I. & Nicol, G. W. Ammonia concentration determines differential growth of ammonia-oxidising archaea and bacteria in soil microcosms. ISME J. 5, 1067–1071 (2011).
Guo, X. et al. Gene-informed decomposition model predicts lower soil carbon loss due to persistent microbial adaptation to warming. Nat. Commun. 11, 4897 (2020).
Frank, D. A. & McNaughton, S. J. Aboveground biomass estimation with the canopy intercept method: a plant growth form caveat. Oikos 57, 57–60 (1990).
Sherry, R. A. et al. Lagged effects of experimental warming and doubled precipitation on annual and seasonal aboveground biomass production in a tallgrass prairie. Glob. Change Biol. 14, 2923–2936 (2008).
Zhou, J. Z. et al. Microbial mediation of carbon-cycle feedbacks to climate warming. Nat. Clim. Change 2, 106–110 (2012).
Li, D., Zhou, X., Wu, L., Zhou, J. & Luo, Y. Contrasting responses of heterotrophic and autotrophic respiration to experimental warming in a winter annual-dominated prairie. Glob. Change Biol. 19, 3553–3564 (2013).
Zhou, J., Bruns, M. A. & Tiedje, J. M. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62, 316–322 (1996).
Wu, L. et al. Phasing amplicon sequencing on Illumina Miseq for robust environmental microbial community analysis. BMC Microbiol. 15, 125 (2015).
Caporaso, J. G. et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624 (2012).
Suzuki, M. T. & Giovannoni, S. J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62, 625–630 (1996).
Takai, K. & Horikoshi, K. Rapid detection and quantification of members of the archaeal community by quantitative PCR using fluorogenic probes. Appl. Environ. Microbiol. 66, 5066–5072 (2000).
Porat, I. et al. Characterization of archaeal community in contaminated and uncontaminated surface stream sediments. Microb. Ecol. 60, 784–795 (2010).
Francis, C. A., Roberts, K. J., Beman, J. M., Santoro, A. E. & Oakley, B. B. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl Acad. Sci. USA 102, 14683–14688 (2005).
Peiffer, J. A. et al. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl Acad. Sci. USA 110, 6548–6553 (2013).
Giardine, B. et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 15, 1451–1455 (2005).
Kong, Y. Btrim: a fast, lightweight adapter and quality trimming program for next-generation sequencing technologies. Genomics 98, 152–153 (2011).
Magoc, T. & Salzberg, S. L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963 (2011).
Edgar, R. C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998 (2013).
Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010).
DeSantis, T. Z. et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072 (2006).
Wang, Q., Garrity, G. M., Tiedje, J. M. & Cole, J. R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267 (2007).
Edgar, R. C. UNOISE2: improved error-correction for Illumina 16S and ITS amplicon sequencing. Preprint available at bioRxiv https://doi.org/10.1101/081257 (2016).
Callahan, B. J. et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016).
Amir, A. et al. Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems 2, e00191-16 (2017).
Kembel, S. W. et al. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26, 1463–1464 (2010).
Sievers, F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011).
Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 5, e9490 (2010).
Munoz, R. et al. Release LTPs104 of the All-Species Living Tree. Syst. Appl. Microbiol. 34, 169–170 (2011).
Oksanen, J. et al. Package ‘vegan’. Community Ecology Package, Version 2.9, 1–295 (The R Project for Statistical Computing, 2013).
Chen, L. X. et al. Comparative metagenomic and metatranscriptomic analyses of microbial communities in acid mine drainage. ISME J. 9, 1579–1592 (2015).
Nakagawa, S. & Schielzeth, H. A general and simple method for obtaining R2 from generalized linear mixed-effects models. Methods Ecol. Evol. 4, 133–142 (2013).
Martiny, J. B., Eisen, J. A., Penn, K., Allison, S. D. & Horner-Devine, M. C. Drivers of bacterial beta-diversity depend on spatial scale. Proc. Natl Acad. Sci. USA 108, 7850–7854 (2011).
Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data (Babraham Bioinformatics, 2010).
Li, W. & Godzik, A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 (2006).
Patel, R. K. & Jain, M. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS ONE 7, e30619 (2012).
Zhou, J. Z. & Ning, D. L. Stochastic community assembly: does it matter in microbial ecology? Microbiol. Mol. Biol. Rev. 81, e00002-17 (2017).
Jaumot, J., Bedia, C. & Tauler, R. Data Analysis for Omic Sciences: Methods and Applications (Elsevier, 2018).
Thevenot, E. A., Roux, A., Xu, Y., Ezan, E. & Junot, C. Analysis of the human adult urinary metabolome variations with age, body mass index, and gender by implementing a comprehensive workflow for univariate and OPLS statistical analyses. J. Proteome Res. 14, 3322–3335 (2015).
Acknowledgements
We thank all former and current members of the Institute for Environmental Genomics for their time and energy in maintaining the long-term climate change experiment. This work was supported by the US Department of Energy, Office of Science, Genomic Science Program under Award Number DE-SC0004601 and DE-SC0010715, and the Office of the Vice President for Research at the University of Oklahoma. The data analysis performed by D.N. and N.X. was also partially supported by NSF Grants EF-2025558 and DEB-2129235.
Author information
Authors and Affiliations
Contributions
All authors contributed intellectual input and assistance to this study. The original concepts were conceived by Y.Z. and J.Z. Field management was carried out by Y.Z., Linwei Wu, M.M.Y., X.Z., X.G., S.J., Z.Y., S.H., J.F., J.K., C.R.C., C.T.B., Y. Fan, J.P.M., Y.O., Y. Fu, D.N., Z.S., N.X., A.Z. and Liyou Wu. Sample collection, soil chemical and microbial characterization were carried out by Y.H., M.M.Y., Linwei Wu, J.G. and Z.G. Data analyses were done by Y.Z. and D.N. with assistance from Linwei Wu and J.Z. All data analysis and integration were guided by J.Z. The manuscript was prepared by Y.Z., D.N., X.L., Y.Y., J.M.T. and J.Z.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Climate Change thanks Federica D’Alò and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Effects of experimental warming on archaeal community composition under unwarmed and warmed conditions at the order level.
Cumulative richness is expressed as the number of operational taxonomic units (OTUs).
Extended Data Fig. 2 The succession of soil archaea communities under unwarmed and warmed treatments by detrended correspondence analysis (DCA).
Individual samples from warmed and unwarmed plots within each year are shown in (a) and the centroids of four replicates from each treatment within each year are shown in (b). The analysis was performed based on Sorensen dissimilarity metric. Warmed samples are clustered together with control samples in year 0 (2009) and separated from control samples in the following seven years (2010–2016).
Extended Data Fig. 3 Temporal changes in community differences between warming and control conditions for archaea and fungi.
The first year is 2009 (year 0). Considering the repeated-measures design, the warming-versus-control dissimilarity values at each block were fitted to the linear mixed-effects (LMMs) models with a fixed effect of time and a random intercept and slope effect among different pairs of plots (blocks). The slopes are presented as a coefficient in fixed effect ± standard error in random effect. The r2 values are calculated (details in Methods), reflecting the variance explained by the whole LMM model. p values were based on permutation tests (two-sided). The lines showed the fixed effects of the LMM.
Extended Data Fig. 4 Temporal changes in community differences between warming and control conditions for orders Nitrososphaerales and Methanomassiliicoccales.
The analysis was performed based on unweighted UniFrac metrics. Considering the repeated-measures design, the warming-versus-control dissimilarity values at each block were fitted to LMMs with a fixed effect of time and a random intercept and slope effect among different pairs of plots (blocks). The slopes are presented as a coefficient in fixed effect ± standard error in random effect. The r2 values are calculated (details in Methods), reflecting the variance explained by the whole LMM model. p values were based on permutation tests (two-sided). The lines showed the fixed effects of the LMM.
Extended Data Fig. 5 Constrained ordination analysis of archaeal communities.
(a) Canonical correspondence analyses (CCA) of soil archaea community and environmental attributes. Tested environmental attributes include soil nitrate (NO3−), ammonium (NH4+), total nitrogen (TN), total organic C (TOC), pH, Precipitation of sampling month (Prcp_SM), temperature, moisutre drought index, C3 and C4 aboveground biomass, plant richness, and total biomass. The insert table shows the significance of each environmental variable in explaining the variations of archaeal community (one-way ANOVA test). (b) CCA-based variation partitioning analysis (VPA) showed the relative proportions of archaeal community variations that can be explained by different types of environmental factors. The numbers within the circles showed the variation explained by each group of environmental factors alone. The numbers between the circles showed the interactions of the two factors on either side and number in the center of the interactions of all three factors.
Extended Data Fig. 6 Constrained ordination analysis of the order Nitrososphaerales.
(a) CCA of the Nitrososphaerales group and environmental attributes. The tested environmental attributes and other properties are the same as in Extended Data Fig. 5 (one-way ANOVA test). Significant tests (P < 0.05) are shown in bold red. (b) CCA-based VPA showed the relative proportions of variations in the Nitrososphaerales group that can be explained by different types of environmental factors. The numbers within the circles showed the variation explained by each group of environmental factors alone. The numbers between the circles showed the interactions of the two factors on either side and number in the center of the interactions of all three factors.
Extended Data Fig. 7 Warming-induced changes of different bins.
Warming-induced difference between warming and control is expressed in percentages for the three dominating ecological processes—homogeneous selection (HoS), dispersal limitation (DL), and drift and others (DR).
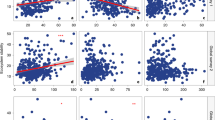
Extended Data Fig. 8 Relationships between archaeal community structure and environmental variables and ecosystem processes under control.
Archaeal community structures, which include taxonomical composition by 16 S rRNA genes and functional gene composition by GeoChip and EcoFUN-MAP, were tested against time, soil and plant variables and ecosystem C fluxes. All the other properties are the same as Fig. 4a.
Supplementary information
Supplementary Information
Supplementary Fig. 1 and Tables 1–9.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, Y., Ning, D., Wu, L. et al. Experimental warming leads to convergent succession of grassland archaeal community. Nat. Clim. Chang. 13, 561–569 (2023). https://doi.org/10.1038/s41558-023-01664-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41558-023-01664-x