Abstract
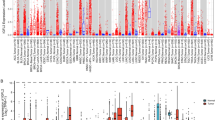
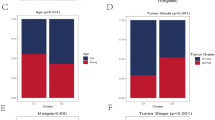
Aging is a complex process that significantly impacts the immune system. The aging-related decline of the immune system, termed immunosenescence, can lead to disease development, including cancer. The perturbation of immunosenescence genes may characterize the associations between cancer and aging. However, the systematical characterization of immunosenescence genes in pan-cancer remains largely unexplored. In this study, we comprehensively investigated the expression of immunosenescence genes and their roles in 26 types of cancer. We developed an integrated computational pipeline to identify and characterize immunosenescence genes in cancer based on the expression profiles of immune genes and clinical information of patients. We identified 2218 immunosenescence genes that were significantly dysregulated in a wide variety of cancers. These immunosenescence genes were divided into six categories based on their relationships with aging. Besides, we assessed the importance of immunosenescence genes in clinical prognosis and identified 1327 genes serving as prognostic markers in cancers. BTN3A1, BTN3A2, CTSD, CYTIP, HIF1AN, and RASGRP1 were associated with ICB immunotherapy response and served as prognostic factors after ICB immunotherapy in melanoma. Collectively, our results furthered the understanding of the relationship between immunosenescence and cancer and provided insights into immunotherapy for patients.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 digital issues and online access to articles
$119.00 per year
only $19.83 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The datasets supporting the conclusions of this article are available in the TCGA repository, (https://tcga-data.nci.nih.gov/) and the GTEx portal (https://gtexportal.org).
References
He S, Sharpless NE. Senescence in health and disease. Cell. 2017;169:1000–11.
Badiola I, Santaolalla F, Garcia-Gallastegui P, Ana SD, Unda F, Ibarretxe G. Biomolecular bases of the senescence process and cancer. A new approach to oncological treatment linked to ageing. Ageing Res Rev. 2015;23:125–38.
Falandry C, Bonnefoy M, Freyer G, Gilson E. Biology of cancer and aging: a complex association with cellular senescence. J Clin Oncol. 2014;32:2604–10.
Lian J, Yue Y, Yu W, Zhang Y. Immunosenescence: a key player in cancer development. J Hematol Oncol. 2020;13:151.
Rodrigues LP, Teixeira VR, Alencar-Silva T, Simonassi-Paiva B, Pereira RW, Pogue R, et al. Hallmarks of aging and immunosenescence: connecting the dots. Cytokine Growth Factor Rev. 2021;59:9–21.
Bulati M, Caruso C, Colonna-Romano G. From lymphopoiesis to plasma cells differentiation, the age-related modifications of B cell compartment are influenced by “inflamm-ageing”. Ageing Res Rev. 2017;36:125–36.
Nikolich-Zugich J. The twilight of immunity: emerging concepts in aging of the immune system. Nat Immunol. 2018;19:10–9.
Gide TN, Wilmott JS, Scolyer RA, Long GV. Primary and acquired resistance to immune checkpoint inhibitors in metastatic melanoma. Clin Cancer Res. 2018;24:1260–70.
Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer. 2019;19:133–50.
Li B, Severson E, Pignon JC, Zhao H, Li T, Novak J, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17:174.
Milling L, Zhang Y, Irvine DJ. Delivering safer immunotherapies for cancer. Adv Drug Deliv Rev. 2017;114:79–101.
Mitnitski A, Collerton J, Martin-Ruiz C, Jagger C, von Zglinicki T, Rockwood K, et al. Age-related frailty and its association with biological markers of ageing. BMC Med. 2015;13:161.
Uyar B, Palmer D, Kowald A, Murua Escobar H, Barrantes I, Moller S, et al. Single-cell analyses of aging, inflammation and senescence. Ageing Res Rev. 2020;64:101156.
Mondal AM, Horikawa I, Pine SR, Fujita K, Morgan KM, Vera E, et al. p53 isoforms regulate aging- and tumor-associated replicative senescence in T lymphocytes. J Clin Invest. 2013;123:5247–57.
Hofmann JW, Zhao X, De Cecco M, Peterson AL, Pagliaroli L, Manivannan J, et al. Reduced expression of MYC increases longevity and enhances healthspan. Cell. 2015;160:477–88.
Gao A, Liu X, Lin W, Wang J, Wang S, Si F, et al. Tumor-derived ILT4 induces T cell senescence and suppresses tumor immunity. J Immunother Cancer. 2021;9:e001536.
Consortium AA. Aging Atlas: a multi-omics database for aging biology. Nucleic Acids Res. 2021;49:D825–D830.
Bhattacharya S, Dunn P, Thomas CG, Smith B, Schaefer H, Chen J, et al. ImmPort, toward repurposing of open access immunological assay data for translational and clinical research. Sci Data. 2018;5:180015.
Breuer K, Foroushani AK, Laird MR, Chen C, Sribnaia A, Lo R, et al. InnateDB: systems biology of innate immunity and beyond–recent updates and continuing curation. Nucleic Acids Res. 2013;41:D1228–1233.
Balliu B, Durrant M, Goede O, Abell N, Li X, Liu B, et al. Genetic regulation of gene expression and splicing during a 10-year period of human aging. Genome Biol. 2019;20:230.
Chatsirisupachai K, Lesluyes T, Paraoan L, Van Loo P, de Magalhaes JP. An integrative analysis of the age-associated multi-omic landscape across cancers. Nat Commun. 2021;12:2345.
Chatsirisupachai K, Palmer D, Ferreira S, de Magalhaes JP. A human tissue-specific transcriptomic analysis reveals a complex relationship between aging, cancer, and cellular senescence. Aging Cell. 2019;18:e13041.
Lu RJ, Taylor S, Contrepois K, Kim M, Bravo JI, Ellenberger M, et al. Multi-omic profiling of primary mouse neutrophils predicts a pattern of sex and age-related functional regulation. Nat Aging. 2021;1:715–33.
Shao T, Wu A, Chen J, Chen H, Lu J, Bai J, et al. Identification of module biomarkers from the dysregulated ceRNA-ceRNA interaction network in lung adenocarcinoma. Mol Biosyst. 2015;11:3048–58.
Li Y, Wang Z, Wang Y, Zhao Z, Zhang J, Lu J, et al. Identification and characterization of lncRNA mediated transcriptional dysregulation dictates lncRNA roles in glioblastoma. Oncotarget. 2016;7:45027–41.
Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–7.
Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28:1747–56.
Garcia-Diaz A, Shin DS, Moreno BH, Saco J, Escuin-Ordinas H, Rodriguez GA, et al. Interferon receptor signaling pathways regulating PD-L1 and PD-L2 expression. Cell Rep. 2017;19:1189–201.
Gide TN, Quek C, Menzies AM, Tasker AT, Shang P, Holst J, et al. Distinct immune cell populations define response to anti-PD-1 Monotherapy and anti-PD-1/Anti-CTLA-4 combined therapy. Cancer Cell. 2019;35:238–255 e236.
Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, et al. Genomic and transcriptomic features of response to Anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165:35–44.
Nathanson T, Ahuja A, Rubinsteyn A, Aksoy BA, Hellmann MD, Miao D, et al. Somatic mutations and neoepitope homology in melanomas treated with CTLA-4 blockade. Cancer Immunol Res. 2017;5:84–91.
Riaz N, Havel JJ, Makarov V, Desrichard A, Urba WJ, Sims JS, et al. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell. 2017;171:934–949 e916.
Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350:207–11.
Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, et al. The immune landscape of cancer. Immunity. 2018;48:812–830 e814.
Donertas HM, Fuentealba Valenzuela M, Partridge L, Thornton JM. Gene expression-based drug repurposing to target aging. Aging Cell. 2018;17:e12819.
Izgi H, Han D, Isildak U, Huang S, Kocabiyik E, Khaitovich P, et al. Inter-tissue convergence of gene expression during ageing suggests age-related loss of tissue and cellular identity. Elife. 2022;11:68048.
Kedlian VR, Donertas HM, Thornton JM. The widespread increase in inter-individual variability of gene expression in the human brain with age. Aging (Albany NY). 2019;11:2253–80.
Barry KC, Hsu J, Broz ML, Cueto FJ, Binnewies M, Combes AJ, et al. A natural killer-dendritic cell axis defines checkpoint therapy-responsive tumor microenvironments. Nat Med. 2018;24:1178–91.
Han J, Weisbrod RM, Shao D, Watanabe Y, Yin X, Bachschmid MM, et al. The redox mechanism for vascular barrier dysfunction associated with metabolic disorders: Glutathionylation of Rac1 in endothelial cells. Redox Biol. 2016;9:306–19.
Miyata K, Imai Y, Hori S, Nishio M, Loo TM, Okada R, et al. Pericentromeric noncoding RNA changes DNA binding of CTCF and inflammatory gene expression in senescence and cancer. Proc Natl Acad Sci USA. 2021;118:35.
Zirkel A, Nikolic M, Sofiadis K, Mallm JP, Brackley CA, Gothe H, et al. HMGB2 loss upon senescence entry disrupts genomic organization and induces CTCF clustering across cell types. Mol Cell. 2018;70:730–744 e736.
Liang Z, Zhao Y, Ruan L, Zhu L, Jin K, Zhuge Q, et al. Impact of aging immune system on neurodegeneration and potential immunotherapies. Prog Neurobiol. 2017;157:2–28.
Bavik C, Coleman I, Dean JP, Knudsen B, Plymate S, Nelson PS. The gene expression program of prostate fibroblast senescence modulates neoplastic epithelial cell proliferation through paracrine mechanisms. Cancer Res. 2006;66:794–802.
Laberge RM, Awad P, Campisi J, Desprez PY. Epithelial-mesenchymal transition induced by senescent fibroblasts. Cancer Microenviron. 2012;5:39–44.
Liu X, Hoft DF, Peng G. Senescent T cells within suppressive tumor microenvironments: emerging target for tumor immunotherapy. J Clin Invest. 2020;130:1073–83.
Parrinello S, Coppe JP, Krtolica A, Campisi J. Stromal-epithelial interactions in aging and cancer: senescent fibroblasts alter epithelial cell differentiation. J Cell Sci. 2005;118:485–96.
Wang T, Notta F, Navab R, Joseph J, Ibrahimov E, Xu J, et al. Senescent carcinoma-associated fibroblasts upregulate IL8 to enhance prometastatic phenotypes. Mol Cancer Res. 2017;15:3–14.
Lanna A, Gomes DC, Muller-Durovic B, McDonnell T, Escors D, Gilroy DW, et al. A sestrin-dependent Erk-Jnk-p38 MAPK activation complex inhibits immunity during aging. Nat Immunol. 2017;18:354–63.
Georgilis A, Klotz S, Hanley CJ, Herranz N, Weirich B, Morancho B, et al. PTBP1-mediated alternative splicing regulates the inflammatory secretome and the pro-tumorigenic effects of senescent cells. Cancer Cell. 2018;34:85–102 e109.
Martincorena I, Campbell PJ. Somatic mutation in cancer and normal cells. Science. 2015;349:1483–9.
Scheffler M, Ihle MA, Hein R, Merkelbach-Bruse S, Scheel AH, Siemanowski J, et al. K-ras mutation subtypes in NSCLC and associated co-occuring mutations in other oncogenic pathways. J Thorac Oncol. 2019;14:606–16.
Braumuller H, Wieder T, Brenner E, Assmann S, Hahn M, Alkhaled M, et al. T-helper-1-cell cytokines drive cancer into senescence. Nature. 2013;494:361–5.
Budovsky A, Craig T, Wang J, Tacutu R, Csordas A, Lourenco J, et al. LongevityMap: a database of human genetic variants associated with longevity. Trends Genet. 2013;29:559–60.
Petti C, Molla A, Vegetti C, Ferrone S, Anichini A, Sensi M. Coexpression of NRASQ61R and BRAFV600E in human melanoma cells activates senescence and increases susceptibility to cell-mediated cytotoxicity. Cancer Res. 2006;66:6503–11.
Raouf S, Weston C, Yucel N. Reproducibility Project: Cancer Biology. Registered report: senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Elife. 2015;4:e04105.
Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419.
Acknowledgements
The authors gratefully thank the TCGA Research Network data repository for providing data for this work.
Funding
This work was supported by the National Natural Science Foundation of China [32070672], Heilongjiang Welfare Fund Organization of disabled Persons [HJ2022-1], and Post doctoral Science Foundation of Heilongjiang Province (LBH-Z19076).
Author information
Authors and Affiliations
Contributions
XFC, SWN and XYW designed the study and performed analysis. SWN and HZ supervised the research and provided critical advice on the study. XYW, SG, PW, HXZ, YS, JG, CYZ and WZ collected data and developed the methodology. SWN and XYW wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, X., Guo, S., Zhou, H. et al. Pan-cancer transcriptomic analysis identified six classes of immunosenescence genes revealed molecular links between aging, immune system and cancer. Genes Immun 24, 81–91 (2023). https://doi.org/10.1038/s41435-023-00197-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41435-023-00197-9