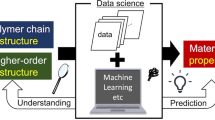
Abstract
This review summarizes recent advances in experimental polymer chemistry supported by data science. The area of polymer informatics is rapidly growing based on cheminformatics, materials informatics, and data science platforms. Data-driven analyses, predictions, and suggestions for experimental polymer research are becoming more practical, and machine learning models can now predict various macromolecular properties with reasonable accuracy. At the same time, the limitations of current polymer informatics are being revealed. Developing appropriate treatments for higher-order structures and experimental procedures is critical to adequately process the hierarchical relationships of polymer systems. Recent attempts to treat this advanced information and future challenges in polymer informatics are discussed.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Ramprasad R, Batra R, Pilania G, Mannodi-Kanakkithodi A, Kim C. Machine learning in materials informatics: recent applications and prospects. Npj Comput Mater. 2017;3:54. https://doi.org/10.1038/s41524-017-0056-5.
Lopez-Bezanilla A, Littlewood PB. Growing field of materials informatics: databases and artificial intelligence. MRS Commun. 2020;10:1–10. https://doi.org/10.1557/mrc.2020.2.
Jackson NE, Webb MA, de Pablo JJ. Recent advances in machine learning towards multiscale soft materials design. Curr Opin Chem Eng. 2019;23:106–14. https://doi.org/10.1016/j.coche.2019.03.005.
Audus DJ, de Pablo JJ. Polymer informatics: opportunities and challenges. ACS Macro Lett. 2017;6:1078–82. https://doi.org/10.1021/acsmacrolett.7b00228.
de Pablo JJ, Jackson NE, Webb MA, Chen L-Q, Moore JE, Morgan D, et al. New frontiers for the materials genome initiative. Npj Comput Mater. 2019;5:41 https://doi.org/10.1038/s41524-019-0173-4.
Kim C, Chandrasekaran A, Huan TD, Das D, Ramprasad R. Polymer genome: a data-powered polymer informatics platform for property predictions. J Phys Chem C. 2018;122:17575–85. https://doi.org/10.1021/acs.jpcc.8b02913.
Mannodi-Kanakkithodi A, Chandrasekaran A, Kim C, Huan TD, Pilania G, Botu V, et al. Scoping the polymer genome: a roadmap for rational polymer dielectrics design and beyond. Mater Today. 2018;21:785–96. https://doi.org/10.1016/j.mattod.2017.11.021.
Amamoto Y. Data-driven approaches for structure-property relationships in polymer science for prediction and understanding. Polym J. 2022;54:957–67. https://doi.org/10.1038/s41428-022-00648-6.
Sha W, Li Y, Tang S, Tian J, Zhao Y, Guo Y, et al. Machine learning in polymer informatics. InfoMat. 2021;3:353–61. https://doi.org/10.1002/inf2.12167.
Chen L, Pilania G, Batra R, Huan TD, Kim C, Kuenneth C, et al. Polymer informatics: current status and critical next steps. Mater Sci Eng R Rep. 2021;144:100595. https://doi.org/10.1016/j.mser.2020.100595.
Schustik SA, Cravero F, Ponzoni I, Díaz MF. Polymer informatics: expert-in-the-loop in QSPR modeling of refractive index. Comput Mater Sci. 2021;194:110460. https://doi.org/10.1016/j.commatsci.2021.110460.
Oaki Y, Igarashi Y. Materials informatics for 2D materials combined with sparse modeling and chemical perspective: toward small-data-driven Chemistry and Materials Science. Bull Chem Soc Jpn. 2021;94:2410–22. https://doi.org/10.1246/bcsj.20210253.
Jordan MI, Mitchell TM. Machine learning: trends, perspectives, and prospects. Science. 2015;349:255–60. https://doi.org/10.1126/science.aaa8415.
Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022;50:D439–44. https://doi.org/10.1093/nar/gkab1061.
Callaway E. ‘It will change everything’: DeepMind’s AI makes gigantic leap in solving protein structures. Nature. 2020;588:203–4. https://doi.org/10.1038/d41586-020-03348-4.
Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–9. https://doi.org/10.1038/s41586-021-03819-2.
Kanda GN, Tsuzuki T, Terada M, Sakai N, Motozawa N, Masuda T, et al. Robotic search for optimal cell culture in regenerative medicine. Elife. 2022;11:e77007. https://doi.org/10.7554/eLife.77007.
Burger B, Maffettone PM, Gusev VV, Aitchison CM, Bai Y, Wang X, et al. A mobile robotic chemist. Nature. 2020;583:237–41. https://doi.org/10.1038/s41586-020-2442-2.
Haven JJ, Baeten E, Claes J, Vandenbergh J, Junkers T. High-throughput polymer screening in microreactors: boosting the Passerini three component reaction. Polym Chem. 2017;8:2972–8. https://doi.org/10.1039/c7py00360a.
Baudis S, Behl M. High-throughput and combinatorial approaches for the development of multifunctional polymers. Macromol Rapid Commun. 2022;43:e2100400. https://doi.org/10.1002/marc.202100400.
Granda JM, Donina L, Dragone V, Long DL, Cronin L. Controlling an organic synthesis robot with machine learning to search for new reactivity. Nature. 2018;559:377–81. https://doi.org/10.1038/s41586-018-0307-8.
Dave A, Mitchell J, Kandasamy K, Wang H, Burke S, Paria B, et al. Autonomous discovery of battery electrolytes with robotic experimentation and machine learning. Cell Rep Phys Sci. 2020;1:100264. https://doi.org/10.1016/j.xcrp.2020.100264.
Shimizu R, Kobayashi S, Watanabe Y, Ando Y, Hitosugi T. Autonomous materials synthesis by machine learning and robotics. APL Mater. 2020;8:111110. https://doi.org/10.1063/5.0020370.
Hatakeyama-Sato K, Tezuka T, Umeki M, Oyaizu K. AI-assisted exploration of superionic Glass-Type Li(+) conductors with aromatic structures. J Am Chem Soc. 2020;142:3301–5. https://doi.org/10.1021/jacs.9b11442.
Hatakeyama-Sato K, Umeki M, Adachi H, Kuwata N, Hasegawa G, Oyaizu K. Exploration of organic superionic glassy conductors by process and materials informatics with lossless graph database. Npj Comput Mater. 2022;8:170. https://doi.org/10.1038/s41524-022-00853-0.
Hatakeyama-Sato K, Tezuka T, Nishikitani Y, Nishide H, Oyaizu K. Synthesis of Lithium-ion conducting polymers designed by machine learning-based prediction and screening. Chem Lett. 2019;48:130–2. https://doi.org/10.1246/cl.180847.
Nagasawa S, Al-Naamani E, Saeki A. Computer-aided screening of conjugated polymers for organic solar cell: classification by random forest. J Phys Chem Lett. 2018;9:2639–46. https://doi.org/10.1021/acs.jpclett.8b00635.
Miyake Y, Saeki A. Machine learning-assisted development of organic solar cell materials: issues, analyses, and outlooks. J Phys Chem Lett. 2021;12:12391–401. https://doi.org/10.1021/acs.jpclett.1c03526.
Komura T, Sakano K, Igarashi Y, Numazawa H, Imai H, Oaki Y. A capacity-prediction model for exploration of organic anodes: discovery of 5-formylsalicylic acid as a high-performance anode active material. ACS Appl Energy Mater. 2022;5:8990–8. https://doi.org/10.1021/acsaem.2c01472.
Numazawa H, Igarashi Y, Sato K, Imai H, Oaki Y. Experiment-oriented materials informatics for efficient exploration of design strategy and new compounds for high-performance organic anode. Adv Theory Simul. 2019;2:1900130. ARTN 190013010.1002/adts.201900130.
Franco AA, Rucci A, Brandell D, Frayret C, Gaberscek M, Jankowski P, et al. Boosting rechargeable batteries R&D by multiscale modeling: myth or reality? Chem Rev. 2019;119:4569–627. https://doi.org/10.1021/acs.chemrev.8b00239.
Pruksawan S, Lambard G, Samitsu S, Sodeyama K, Naito M. Prediction and optimization of epoxy adhesive strength from a small dataset through active learning. Sci Technol Adv Mater. 2019;20:1010–21. https://doi.org/10.1080/14686996.2019.1673670.
Yamada H, Liu C, Wu S, Koyama Y, Ju S, Shiomi J, et al. Predicting materials properties with little data using shotgun transfer learning. ACS Cent Sci 2019;5:1717–30. https://doi.org/10.1021/acscentsci.9b00804.
Wu S, Kondo Y, Kakimoto M-A, Yang B, Yamada H, Kuwajima I, et al. Machine-learning-assisted discovery of polymers with high thermal conductivity using a molecular design algorithm. Npj Comput Mater. 2019;5:66. https://doi.org/10.1038/s41524-019-0203-2.
Taniwaki H, Kaneko H. Molecular design of monomers by considering the dielectric constant and stability of the polymer. Polym Eng Sci. 2022;62:2750–6. https://doi.org/10.1002/pen.26058.
Hastie T, Tibshirani R, Friedman J, The Elements of Statistical Learning, Springer New York, NY, 2009 https://doi.org/10.1007/978-0-387-84858-7.
David L, Thakkar A, Mercado R, Engkvist O. Molecular representations in AI-driven drug discovery: a review and practical guide. J Cheminform 2020;12:56. https://doi.org/10.1186/s13321-020-00460-5.
Chen Y, Kirchmair J. Cheminformatics in natural product-based drug Discovery. Mol Inf. 2020;39:e2000171. https://doi.org/10.1002/minf.202000171.
Muratov EN, Bajorath J, Sheridan RP, Tetko IV, Filimonov D, Poroikov V, et al. Tropsha QSAR without borders. Chem Soc Rev. 2020;49:3525–64. https://doi.org/10.1039/d0cs00098a.
Papers With Code: The latest in Machine Learning. https://paperswithcode.com/datasets.
Lin TS, Coley CW, Mochigase H, Beech HK, Wang W, Wang Z, et al. BigSMILES: a structurally-based line notation for describing macromolecules. ACS Cent Sci. 2019;5:1523–31. https://doi.org/10.1021/acscentsci.9b00476.
Sharma A, Kumar R, Ranjta S, Varadwaj PK. SMILES to smell: decoding the structure-odor relationship of Chemical Compounds Using the Deep Neural Network Approach. J. Chem. Inf. Model. 2021. https://doi.org/10.1021/acs.jcim.0c01288.
Drefahl A. CurlySMILES: a chemical language to customize and annotate encodings of molecular and nanodevice structures. J Cheminform. 2011;3:1 https://doi.org/10.1186/1758-2946-3-1.
Kuenneth C, Rajan AC, Tran H, Chen L, Kim C, Ramprasad R. Polymer informatics with multi-task learning. Patterns (N. Y). 2021;2:100238 https://doi.org/10.1016/j.patter.2021.100238.
Hatakeyama-Sato K, Oyaizu K. Integrating multiple materials science projects in a single neural network. Commun Mater 2020;1:49 https://doi.org/10.1038/s43246-020-00052-8. article number
Shields BJ, Stevens J, Li J, Parasram M, Damani F, Alvarado JIM, et al. Bayesian reaction optimization as a tool for chemical synthesis. Nature. 2021;590:89–96. https://doi.org/10.1038/s41586-021-03213-y.
Bilal M, Oyedele LO, Qadir J, Munir K, Ajayi SO, Akinade OO, et al. Big Data in the construction industry: a review of present status, opportunities, and future trends. Adv Eng Inform. 2016;30:500–21. https://doi.org/10.1016/j.aei.2016.07.001.
Dong S, Wang P, Abbas K. A survey on deep learning and its applications. Comput Sci Rev. 2021;40:100379. https://doi.org/10.1016/j.cosrev.2021.100379.
Janiesch C, Zschech P, Heinrich K. Machine learning and deep learning. Electron Mark. 2021;31:685–95. https://doi.org/10.1007/s12525-021-00475-2.
Sepehri B. A review on created QSPR models for predicting ionic liquids properties and their reliability from chemometric point of view. J Mol Liq. 2020;297:112013. https://doi.org/10.1016/j.molliq.2019.112013.
Curtarolo S, Hart GL, Nardelli MB, Mingo N, Sanvito S, Levy O. The high-throughput highway to computational materials design. Nat Mater. 2013;12:191–201. https://doi.org/10.1038/nmat3568.
Matsubara M, Suzumura A, Ohba N, Asahi R. Identifying superionic conductors by materials informatics and high-throughput synthesis. Commun Mater. 2020;1:5 https://doi.org/10.1038/s43246-019-0004-7.
Wang Y, Richards WD, Ong SP, Miara LJ, Kim JC, Mo Y, et al. Design principles for solid-state lithium superionic conductors. Nat Mater. 2015;14:1026–31. https://doi.org/10.1038/nmat4369.
Otsuka S, Kuwajima I, Hosoya J, Xu Y, Yamazaki M. PoLyInfo: Polymer database for polymeric materials design. 2011 International Conference on Emerging Intelligent Data and Web Technologies 2011:22–29. https://doi.org/10.1109/eidwt.2011.13.
Mark J, Polymer data handbook, Oxford University Press, New York 1998.
Mark J, Physical Properties of Polymers Handbook, Springer, New York 2006.
Luo Y, Bag S, Zaremba O, Cierpka A, Andreo J, Wuttke S, et al. MOF synthesis prediction enabled by automatic data mining and machine learning. Angew. Chem Int Ed. 2022;61:e202200242. https://doi.org/10.1002/anie.202200242.
Wakiya T, Kamakura Y, Shibahara H, Ogasawara K, Saeki A, Nishikubo R, et al. Machine-learning-assisted selective synthesis of a semiconductive silver thiolate coordination polymer with segregated paths for holes and electrons. Angew Chem Int Ed. 2021;60:23217–24. https://doi.org/10.1002/anie.202110629.
Nickel M, Murphy K, Tresp V, Gabrilovich E. A review of relational machine learning for knowledge graphs. Proc IEEE Inst Electr Electron Eng. 2016;104:11–33. https://doi.org/10.1109/jproc.2015.2483592.
Qiao L, Zhang L, Chen S, Shen D. Data-driven graph construction and graph learning: a review. Neurocomputing. 2018;312:336–51. https://doi.org/10.1016/j.neucom.2018.05.084.
Gaudelet T, Day B, Jamasb AR, Soman J, Regep C, Liu G, et al. Utilizing graph machine learning within drug discovery and development. Brief Bioinform. 2021;22:1. https://doi.org/10.1093/bib/bbab159.
Wang X, Zhu W. in Proceedings of the 27th ACM SIGKDD Conference on Knowledge Discovery & Data Mining, 2021, 4082–3.
Mrdjenovich D, Horton MK, Montoya JH, Legaspi CM, Dwaraknath S, Tshitoyan V, et al. propnet: a knowledge graph for Materials Science. Matter. 2020;2:464–80. https://doi.org/10.1016/j.matt.2019.11.013.
Wieder O, Kohlbacher S, Kuenemann M, Garon A, Ducrot P, Seidel T, et al. A compact review of molecular property prediction with graph neural networks. Drug Disco Today Technol. 2020;37:1–12. https://doi.org/10.1016/j.ddtec.2020.11.009.
Jiang D, Wu Z, Hsieh CY, Chen G, Liao B, Wang Z, et al. Could graph neural networks learn better molecular representation for drug discovery? A comparison study of descriptor-based and graph-based models. J Cheminform. 2021;13:12. https://doi.org/10.1186/s13321-020-00479-8.
Walsh E, Cho I. Using Evernote as an electronic lab notebook in a translational science laboratory. J Lab Autom. 2013;18:229–34. https://doi.org/10.1177/2211068212471834.
Tremouilhac P, Nguyen A, Huang YC, Kotov S, Lutjohann DS, Hubsch F, et al. Chemotion ELN: an Open Source electronic lab notebook for chemists in academia. J Cheminform. 2017;9:54. https://doi.org/10.1186/s13321-017-0240-0.
Ghiandoni GM, Bodkin MJ, Chen B, Hristozov D, Wallace JEA, Webster J, et al. Development and application of a data-driven reaction classification model: comparison of an electronic lab notebook and medicinal Chemistry Literature. J Chem Inf Model. 2019;59:4167–87. https://doi.org/10.1021/acs.jcim.9b00537.
Bhowmik R, Sihn S, Pachter R, Vernon JP. Prediction of the specific heat of polymers from experimental data and machine learning methods. Polymer. 2021;220:123558. https://doi.org/10.1016/j.polymer.2021.123558.
Yang J, Tao L, He J, McCutcheon JR, Li Y. Machine learning enables interpretable discovery of innovative polymers for gas separation membranes. Sci Adv. 2022;8:eabn9545. https://doi.org/10.1126/sciadv.abn9545.
Liang Z, Li Z, Zhou S, Sun Y, Yuan J, Zhang C. Machine-learning exploration of polymer compatibility. Cell Rep. Phys Sci. 2022;3:100931. https://doi.org/10.1016/j.xcrp.2022.100931.
Park J, Shim Y, Lee F, Rammohan A, Goyal S, Shim M, et al. Prediction and interpretation of polymer properties using the graph convolutional network. ACS Polym Au. 2022;2:213–22. https://doi.org/10.1021/acspolymersau.1c00050.
Li D. The MNIST database of handwritten digit images for machine learning research [Best of the Web]. IEEE Signal Process Mag. 2012;29:141–2. https://doi.org/10.1109/msp.2012.2211477.
Zardecki C, Dutta S, Goodsell DS, Lowe R, Voigt M, Burley SK. PDB-101: Educational resources supporting molecular explorations through biology and medicine. Protein Sci. 2022;31:129–40. https://doi.org/10.1002/pro.4200.
Wu Z, Ramsundar B, Feinberg EN, Gomes J, Geniesse C, Pappu AS, et al. MoleculeNet: a benchmark for molecular machine learning. Chem Sci. 2018;9:513–30. https://doi.org/10.1039/c7sc02664a.
Levin I. NIST Inorganic Crystal Structure Database (ICSD), National Institute of Standards and Technology. https://doi.org/10.18434/M32147.
Vrandečić D, Krötzsch Wikidata M. Commun. ACM. 2014;57:78–85. https://doi.org/10.1145/2629489.
Katsura Y, Kumagai M, Kodani T, Kaneshige M, Ando Y, Gunji S, et al. Data-driven analysis of electron relaxation times in PbTe-type thermoelectric materials. Sci Technol Adv Mater. 2019;20:511–20. https://doi.org/10.1080/14686996.2019.1603885.
Duchowicz PR, Fioressi SE, Bacelo DE, Saavedra LM, Toropova AP, Toropov AA. QSPR studies on refractive indices of structurally heterogeneous polymers. Chemom Intell Lab Syst. 2015;140:86–91. https://doi.org/10.1016/j.chemolab.2014.11.008.
Gedeck P, Rohde B, Bartels C. QSAR-how good is it in practice? Comparison of descriptor sets on an unbiased cross section of corporate data sets. J Chem Inf Model. 2006;46:1924–36. https://doi.org/10.1021/ci050413p.
Dam HC, Nguyen VC, Pham TL, Nguyen AT, Terakura K, Miyake T, et al. Important descriptors and descriptor groups of curie temperatures of rare-earth transition-metal binary alloys. J Phys Soc Jpn. 2018;87:113801. https://doi.org/10.7566/jpsj.87.113801.
Rogers D, Hahn M. Extended-connectivity fingerprints. J Chem Inf Model. 2010;50:742–54. https://doi.org/10.1021/ci100050t.
Capecchi A, Probst D, Reymond JL. One molecular fingerprint to rule them all: drugs, biomolecules, and the metabolome. J Cheminform. 2020;12:43. https://doi.org/10.1186/s13321-020-00445-4.
Lambard G, Gracheva E. SMILES-X: autonomous molecular compounds characterization for small datasets without descriptors. Mach Llearn: Sci Technol. 2020;1:025004. https://doi.org/10.1088/2632-2153/ab57f3.
Zhou J, Cui G, Zhang Z, Yang C, Liu Z, Wang L, et al. Graph neural networks: a review of methods and applications. 2018;arXiv:1812.08434. arXiv:1812.08434.
Duvenaudy D, Maclauriny D, Aguilera-Iparraguirre J, Gomez-Bombarelli R, Hirzel T, Aspuru-Guzik A, et al. Convolutional networks on graphs for learning molecular fingerprints. 2015;arXiv:1509.09292. https://doi.org/10.48550/arXiv.1509.09292.
Zhong S, Hu J, Yu X, Zhang H. Molecular image-convolutional neural network (CNN) assisted QSAR models for predicting contaminant reactivity toward OH radicals: Transfer learning, data augmentation and model interpretation. Chem Eng J. 2021;408:127998. https://doi.org/10.1016/j.cej.2020.127998.
Gomez-Bombarelli R, Wei JN, Duvenaud D, Hernandez-Lobato JM, Sanchez-Lengeling B, Sheberla D, et al. Automatic Chemical design using a data-driven continuous representation of molecules. ACS Cent Sci. 2018;4:268–76. https://doi.org/10.1021/acscentsci.7b00572.
Shimizu Y, Kurokawa T, Arai H, Washizu H. Higher-order structure of polymer melt described by persistent homology. Sci Rep. 2021;11:2274. https://doi.org/10.1038/s41598-021-80975-5.
Buchet M, Hiraoka Y, Obayashi I. Persistent Homology and Materials Informatics. In: Tanaka I (eds) Nanoinformatics, Springer, Singapore 2018. https://doi.org/10.1007/978-981-10-7617-6_5.
Maric M, Marano J, Cody RB, Bridge C. DART-MS: a new analytical technique for forensic paint analysis. Anal Chem. 2018;90:6877–84. https://doi.org/10.1021/acs.analchem.8b01067.
Cody RB, Fouquet TNJ, Takei C. Thermal desorption and pyrolysis direct analysis in real time mass spectrometry for qualitative characterization of polymers and polymer additives. Rapid Commun Mass Spectrom. 2020;34:e8687. https://doi.org/10.1002/rcm.8687.
Stafford CM, Guo S, Harrison C, Chiang MYM. Combinatorial and high-throughput measurements of the modulus of thin polymer films. Rev Sci Instrum. 2005;76:062207. https://doi.org/10.1063/1.1906085.
Frenklach M. Transforming data into knowledge—process Informatics for combustion chemistry. Proc Combust Inst. 2007;31:125–40. https://doi.org/10.1016/j.proci.2006.08.121.
Yamakage S, Kaneko H. Design of adaptive soft sensor based on Bayesian optimization. Case Stud Chem Env Eng. 2022;6:100237. https://doi.org/10.1016/j.cscee.2022.100237.
Nagy ZK, Braatz RD. Advances and new directions in crystallization control. Annu Rev Chem Biomol Eng. 2012;3:55–75. https://doi.org/10.1146/annurev-chembioeng-062011-081043.
Bruckstein AM, Donoho DL, Elad M. From sparse solutions of systems of equations to sparse modeling of signals and images. SIAM Rev. 2009;51:34–81. https://doi.org/10.1137/060657704.
Udrescu SM, Tegmark M. AI Feynman: a physics-inspired method for symbolic regression. Sci Adv. 2020;6:eaay2631. https://doi.org/10.1126/sciadv.aay2631.
Iwasaki Y, Ishida M. Data-driven formulation of natural laws by recursive-LASSO-based symbolic regression. arXiv. 2021. https://doi.org/10.48550/arXiv.2102.09210.
Gillies S. Shapely: manipulation and analysis of geometric objects. 2007. https://github.com/Toblerity/Shapely.
Molnar C. Interpretable machine learning: a guide for making black box models explainable.
Hatakeyama-Sato K, Igarashi Y, Kashikawa T, Kimura K, Oyaizu K. Quantum circuit learning to predict experimental chemical properties. ChemRxiv. 2022. https://doi.org/10.26434/chemrxiv-2022-cz7wr-v2.
Mitarai K, Negoro M, Kitagawa M, Fujii K. Quantum circuit learning. Phys Rev A. 2018;98:032309. ARTN 03230910.1103/PhysRevA.98.032309.
Suzuki T, Katouda M. Predicting toxicity by quantum machine learning. J Phys Commun. 2020;4:125012. https://doi.org/10.1088/2399-6528/abd3d8.
Kishino M, Matsumoto K, Kobayashi Y, Taguchi R, Akamatsu N, Shishido A. Fatigue life prediction of bending polymer films using random forest. Int J Fatigue. 2023;166:107230. https://doi.org/10.1016/j.ijfatigue.2022.107230.
Sahu H, Li H, Chen L, Rajan AC, Kim C, Stingelin N, et al. An informatics approach for designing conducting polymers. ACS Appl Mater Interfaces. 2021;13:53314–22. https://doi.org/10.1021/acsami.1c04017.
Shahriari B, Swersky K, Wang Z, Adams RP, de Freitas N. Taking the human out of the loop: a review of Bayesian optimization. Proc IEEE Inst Electr Electron Eng. 2016;104:148–75. https://doi.org/10.1109/jproc.2015.2494218.
Shimizu N, Kaneko H. Direct inverse analysis based on Gaussian mixture regression for multiple objective variables in material design. Mater Des. 2020;196:109168. https://doi.org/10.1016/j.matdes.2020.109168.
Haruna SI, Zhu H, Umar IK, Shao J, Adamu M, Ibrahim YE. Gaussian process regression model for the prediction of the compressive strength of polyurethane-based polymer concrete for runway repair: a comparative approach. IOP Conf Ser: Earth Environ Sci. 2022;1026:012007. https://doi.org/10.1088/1755-1315/1026/1/012007.
Kuzminykh D, Polykovskiy D, Kadurin A, Zhebrak A, Baskov I, Nikolenko S, et al. 3D molecular representations based on the wave transform for convolutional neural networks. Mol Pharm. 2018;15:4378–85. https://doi.org/10.1021/acs.molpharmaceut.7b01134.
Hatakeyama-Sato K, Adachi H, Umeki M, Kashikawa T, Kimura K, Oyaizu K. Automated design of Li(+) -conducting polymer by quantum-inspired annealing. Macromol Rapid Commun. 2022:e2200385. https://doi.org/10.1002/marc.202200385.
Hatakeyama-Sato K, Oyaizu K. Generative models for extrapolation prediction in materials informatics. ACS Omega. 2021;6:14566–74. https://doi.org/10.1021/acsomega.1c01716.
Gao M, Zhang J, Yu J, Li J, Wen J, Xiong Q. Recommender systems based on generative adversarial networks: a problem-driven perspective. Inf Sci. 2021;546:1166–85. https://doi.org/10.1016/j.ins.2020.09.013.
Hatakeyama-Sato K: ion_predictor. https://github.com/KanHatakeyama/ion_predictor.
Bunn CW. The melting points of chain polymers. J Polym Sci. 1955;16:323–43. https://doi.org/10.1002/pol.1955.120168222.
Ogden S, Klintberg L, Thornell G, Hjort K, Bodén R. Review on miniaturized paraffin phase change actuators, valves, and pumps. Microfluid Nanofluidics. 2013;17:53–71. https://doi.org/10.1007/s10404-013-1289-3.
Watanabe S. Knowing and Guessing: A Quantitative Study of Inference and Information, Wiley, New York 1969 https://archive.org/details/knowingguessingq0000wata.
Sutton C, Boley M, Ghiringhelli LM, Rupp M, Vreeken J, Scheffler M. Identifying domains of applicability of machine learning models for materials science. Nat Commun. 2020;11:4428. https://doi.org/10.1038/s41467-020-17112-9.
Kaneko H. Data visualization, regression, applicability domains and inverse analysis based on generative topographic mapping. Mol Inform. 2019;38:e1800088. https://doi.org/10.1002/minf.201800088.
Shen K-H, Fan M, Hall LM. Molecular dynamics simulations of ion-containing polymers using generic coarse-grained models. Macromolecules. 2021;54:2031–52. https://doi.org/10.1021/acs.macromol.0c02557.
Xie T, France-Lanord A, Wang Y, Lopez J, Stolberg MA, Hill M, et al. Accelerating amorphous polymer electrolyte screening by learning to reduce errors in molecular dynamics simulated properties. Nat Commun. 2022;13:3415. https://doi.org/10.1038/s41467-022-30994-1.
Doerr S, Harvey MJ, Noe F, De Fabritiis G. HTMD: high-throughput molecular dynamics for molecular discovery. J Chem Theory Comput. 2016;12:1845–52. https://doi.org/10.1021/acs.jctc.6b00049.
Takamoto S, Shinagawa C, Motoki D, Nakago K, Li W, Kurata I, et al. Towards universal neural network potential for material discovery applicable to arbitrary combination of 45 elements. Nat Commun. 2022;13:2991. https://doi.org/10.1038/s41467-022-30687-9.
Polymer Property Predictor and Database. https://pppdb.uchicago.edu/.
Polymer database (CROW). https://www.polymerdatabase.com/.
Polymers: A Property Database https://poly.chemnetbase.com/faces/polymers/PolymerSearch.xhtml.
Alesadi A, Cao Z, Li Z, Zhang S, Zhao H, Gu X, et al. Machine learning prediction of glass transition temperature of conjugated polymers from chemical structure. Cell Rep Phys Sci. 2022;3:100911. https://doi.org/10.1016/j.xcrp.2022.100911.
Funding
This work was partially supported by Grants-in-Aid for Scientific Research (Grant Nos. 22H04623 and 21H02017) from MEXT, Japan, and the JST FOREST Program (Grant No. JPMJFR213V).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The author declares no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hatakeyama-Sato, K. Recent advances and challenges in experiment-oriented polymer informatics. Polym J 55, 117–131 (2023). https://doi.org/10.1038/s41428-022-00734-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41428-022-00734-9
This article is cited by
-
Advances in materials informatics: a review
Journal of Materials Science (2024)
-
Artificial intelligence driven design of catalysts and materials for ring opening polymerization using a domain-specific language
Nature Communications (2023)