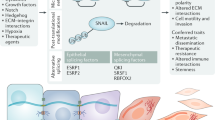
Abstract
The epigenome coordinates spatial-temporal specific gene expression during development and in adulthood, for the maintenance of homeostasis and upon tissue repair. The upheaval of the epigenetic landscape is a key event in the onset of many pathologies including tumours, where epigenetic changes cooperate with genetic aberrations to establish the neoplastic phenotype and to drive cell plasticity during its evolution. DNA methylation, histone modifiers and readers or other chromatin components are indeed often altered in cancers, such as carcinomas that develop in epithelia. Lining the surfaces and the cavities of our body and acting as a barrier from the environment, epithelia are frequently subjected to acute or chronic tissue damages, such as mechanical injuries or inflammatory episodes. These events can activate plasticity mechanisms, with a deep impact on cells’ epigenome. Despite being very effective, tissue repair mechanisms are closely associated with tumour onset. Here we review the similarities between tissue repair and carcinogenesis, with a special focus on the epigenetic mechanisms activated by cells during repair and opted by carcinoma cells in multiple epithelia. Moreover, we discuss the recent findings on inflammatory and wound memory in epithelia and describe the epigenetic modifications that characterise them. Finally, as wound memory in epithelial cells promotes carcinogenesis, we highlight how it represents an early step for the establishment of field cancerization.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Rivera CM, Ren B. Mapping Human Epigenomes. Cell 2013;155:39–55.
Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science 2010;330:612–6.
Waddington CH. The epigenotype. Int J Epidemiol. 2012;41:10–3.
Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell 2007;128:669–81.
Cheng Y, He C, Wang M, Ma X, Mo F, Yang S, et al. Targeting epigenetic regulators for cancer therapy: mechanisms and advances in clinical trials. Signal Transduct Target Ther. 2019;4:1–39.
Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17:487–500.
Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8:253–62.
Mohammad HP, Barbash O, Creasy CL. Targeting epigenetic modifications in cancer therapy: erasing the roadmap to cancer. Nat Med. 2019;25:403–18.
Flavahan WA, Gaskell E, Bernstein BE. Epigenetic plasticity and the hallmarks of cancer. Science. 2017;357:eaal2380.
Audia JE, Campbell RM. Histone modifications and cancer. Cold Spring Harb Perspect Biol. 2016;8:a019521.
Donati G, Watt FM. Stem cell heterogeneity and plasticity in epithelia. Cell Stem Cell. 2015;16:465–76.
Blanpain C, Fuchs E. Plasticity of epithelial stem cells in tissue regeneration. Science 2014;344:1242281.
Adkins-Threats M, Mills JC. Cell plasticity in regeneration in the stomach and beyond. Curr Opin Genet Dev. 2022;75:101948.
Sun X, Joost S, Kasper M. Plasticity of epithelial cells during skin wound healing. Cold Spring Harb Perspect Biol. 2023;15:a041232.
Katsuyama T, Paro R. Epigenetic reprogramming during tissue regeneration. FEBS Lett. 2011;585:1617–24.
Yu H, Wang Y, Wang D, Yi Y, Liu Z, Wu M, et al. Landscape of the epigenetic regulation in wound healing. Front Physiol. 2022;13:949498.
Flier JS, Underhill LH, Dvorak HF. Tumors: wounds that do not heal. Similarities tumor Strom Gener wound healing N. Engl J Med. 1986;315:1650–9.
Arwert EN, Hoste E, Watt FM. Epithelial stem cells, wound healing and cancer. Nat Rev Cancer. 2012;12:170–80.
Ge Y, Fuchs E. Stretching the limits: From homeostasis to stem cell plasticity in wound healing and cancer. Nat Rev Genet. 2018;19:311–25.
MacCarthy-Morrogh L, Martin P. The hallmarks of cancer are also the hallmarks of wound healing. Sci Signal. 2020;13:eaay8690.
Kang S, Chovatiya G, Tumbar T. Epigenetic control in skin development, homeostasis and injury repair. Exp Dermatol. 2019;28:453–63.
Shibata S. Chromatin dynamics and epigenetics in skin stress adaptation. J Dermatol Sci. 2021;103:66–72.
Noodleman FR, Pollack SV. Trauma as a possible etiologic factor in basal cell carcinoma. J Dermatol Surg Oncol. 1986;12:841–6.
Özyazgan I, Kontaş O. Previous injuries or scars as risk factors for the development of basal cell carcinoma. Scand J Plast Reconstr Surg Hand Surg. 2004;38:11–5.
Hartnett L, Egan LJ. Inflammation, DNA methylation and colitis-associated cancer. Carcinogenesis 2012;33:723–31.
Valenzuela MA, Canales J, Corvalán AH, Quest AFG. Helicobacter pylori -induced inflammation and epigenetic changes during gastric carcinogenesis. World J Gastroenterol WJG Press. 2015;21:12742–56.
Ruggiero P. Helicobacter pylori and inflammation. Curr Pharm Des. 2010;16:4225–36.
Walter ND, Rice PL, Redente EF, Kauvar EF, Lemond L, Aly T, et al. Wound healing after trauma may predispose to lung cancer metastasis: review of potential mechanisms. Am J Respir Cell Mol Biol. 2011;44:591–6.
Curtius K, Wright NA, Graham TA. An evolutionary perspective on field cancerization. Nat Rev Cancer. 2018;18:19–32.
Jassim A, Rahrmann EP, Simons BD, Gilbertson RJ. Cancers make their own luck: theories of cancer origins. Nat Rev Cancer. 2023;23:710–24.
Ordovas-Montanes J, Beyaz S, Rakoff-Nahoum S, Shalek AK. Distribution and storage of inflammatory memory in barrier tissues. Nat Rev Immunol. 2020;20:308–20.
Naik S, Fuchs E. Inflammatory memory and tissue adaptation in sickness and in health. Nature 2022;607:249–55.
Del Poggetto E, Ho IL, Balestrieri C, Yen EY, Zhang S, Citron F, et al. Epithelial memory of inflammation limits tissue damage while promoting pancreatic tumorigenesis. Science 2021;373:eabj0486.
Falvo DJ, Grimont A, Zumbo P, Yang JL, Osterhoudt A, Pan G, et al. An epigenetic memory of inflammation controls context-dependent lineage plasticity in the pancreas. bioRxiv. 2022;2021.11.01.466807.
Alonso-Curbelo D, Ho YJ, Burdziak C, Maag JLV, Morris JP, Chandwani R, et al. A gene–environment-induced epigenetic program initiates tumorigenesis. Nature 2021;590:642–8.
Levra Levron C, Watanabe M, Proserpio V, Piacenti G, Lauria A, Kaltenbach S, et al. Tissue memory relies on stem cell priming in distal undamaged areas. Nat Cell Biol. 2023;25:740–53.
Dvorak HF. Tumors: Wounds that do not heal-Redux. Cancer. Immunol Res. 2015;3:1.
Guan Y, Yang YJ, Nagarajan P, Ge Y. Transcriptional and signalling regulation of skin epithelial stem cells in homeostasis, wounds and cancer. Exp Dermatol. 2021;30:529–45.
Lambert AW, Weinberg RA, Linking EMT. programmes to normal and neoplastic epithelial stem cells. Nat Rev Cancer. 2021;21:325–38.
Leopold PL, Vincent J, Wang H. A comparison of epithelial-to-mesenchymal transition and re-epithelialization. Semin Cancer Biol. 2012;22:471–83.
Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med. 2013;19:1438–49.
Singh K, Rustagi Y, Abouhashem AS, Tabasum S, Verma P, Hernandez E, et al. Genome-wide DNA hypermethylation opposes healing in patients with chronic wounds by impairing epithelial-mesenchymal transition. J Clin Invest. 2022;132:e157279.
Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–32.
Shiraishi T, Verdone JE, Huang J, Kahlert UD, Hernandez JR, Torga G, et al. Glycolysis is the primary bioenergetic pathway for cell motility and cytoskeletal remodeling in human prostate and breast cancer cells. Oncotarget 2015;6:130–43.
Konieczny P, Xing Y, Sidhu I, Subudhi I, Mansfield KP, Hsieh B, et al. Interleukin-17 governs hypoxic adaptation of injured epithelium. Science 2022;377:eabg9302.
Iglesias-Bartolome R, Uchiyama A, Molinolo AA, Abusleme L, Brooks SR, Callejas-Valera JL, et al. Transcriptional signature primes human oral mucosa for rapid wound healing. Sci Transl Med. 2018;10:eaap8798.
Siegle JM, Basin A, Sastre-Perona A, Yonekubo Y, Brown J, Sennett R, et al. SOX2 is a cancer-specific regulator of tumour initiating potential in cutaneous squamous cell carcinoma. Nat Commun. 2014;5:4511.
Ferone G, Song JY, Sutherland KD, Bhaskaran R, Monkhorst K, Lambooij JP, et al. SOX2 Is the Determining Oncogenic Switch in Promoting Lung Squamous Cell Carcinoma from Different Cells of Origin. Cancer Cell. 2016;30:519.
Wu Z, Zhou J, Zhang X, Zhang Z, Xie Y, Liu Jbin, et al. Reprogramming of the esophageal squamous carcinoma epigenome by SOX2 promotes ADAR1 dependence. Nat Genet. 2021;53:881–94.
Cangkrama M, Wietecha M, Werner S. Wound repair, scar formation, and cancer: converging on activin trends in molecular medicine. Trends Mol Med. 2020;26:1107–17.
Schäfer M, Werner S. Cancer as an overhealing wound: an old hypothesis revisited. Nat Rev Mol Cell Biol. 2008;9:628–38.
Lafave LM, Savage RE, Buenrostro JD. Single-Cell Epigenomics Reveals Mechanisms of Cancer Progression. Annu Rev Cancer Biol. 2022;6:167–85.
Della Chiara G, Gervasoni F, Fakiola M, Godano C, D’Oria C, Azzolin L, et al. Epigenomic landscape of human colorectal cancer unveils an aberrant core of pan-cancer enhancers orchestrated by YAP/TAZ. Nature. Communications 2021;12:1–18.
Hanahan D, Weinberg RA. The Hallmarks of Cancer. Cell. 2000;100:57–70.
Ge Y, Gomez NC, Adam RC, Yuan S, Elemento O, Fuchs E. Stem cell lineage infidelity drives wound repair and cancer. Cell. 2017;169:636–50.
Vezzani B, Carinci M, Previati M, Giacovazzi S, Della Sala M, Gafà R, et al. Epigenetic Regulation: A Link between Inflammation and Carcinogenesis. Cancers MDPI. 2022;14:1221.
Kang S, Long K, Wang S, Sada A, Tumbar T. Histone H3 K4/9/27 Trimethylation Levels Affect Wound Healing and Stem Cell Dynamics in Adult Skin. Stem Cell Rep. 2020;14:34–48.
Grinat J, Heuberger J, Vidal RO, Goveas N, Kosel F, Berenguer-Llergo A, et al. The epigenetic regulator Mll1 is required for Wnt-driven intestinal tumorigenesis and cancer stemness. Nat Commun. 2020;11:6422.
Kimball AS, Joshi A, Carson WF, Boniakowski AE, Schaller M, Allen R, et al. The Histone Methyltransferase MLL1 Directs Macrophage-Mediated Inflammation in Wound Healing and Is Altered in a Murine Model of Obesity and Type 2 Diabetes. Diabetes 2017;66:2459–71.
Chen R, Zhao WQ, Fang C, Yang X, Ji M. Histone methyltransferase SETD2: a potential tumor suppressor in solid cancers. J Cancer. 2020;11:3349.
Li X, Liu C, Zhu Y, Rao H, Liu M, Gui L, et al. SETD2 epidermal deficiency promotes cutaneous wound healing via activation of AKT/mTOR Signalling. Cell Prolif. 2021;54:e13045.
Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su I-hsin, Hannon G. et al. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136:1122–35.
Chiacchiera F, Rossi A, Jammula S, Zanotti M, Pasini D. PRC2 preserves intestinal progenitors and restricts secretory lineage commitment. EMBO J. 2016;35:2301–14.
Shaw T, Martin P. Epigenetic reprogramming during wound healing: loss of polycomb-mediated silencing may enable upregulation of repair genes. EMBO Rep. 2009;10:881–6.
Comet I, Riising EM, Leblanc B, Helin K. Maintaining cell identity: PRC2-mediated regulation of transcription and cancer. Nat Rev Cancer. 2016;16:803–10.
Chang CJ, Yang JY, Xia W, Chen CT, Xie X, Chao CH. et al. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-β-catenin signaling. Cancer Cell. 2011;19:86–100.
Xue C, Wang K, Jiang X, Gu C, Yu G, Zhong Y, et al. The Down-Regulation of SUZ12 Accelerates the Migration and Invasion of Liver Cancer Cells via Activating ERK1/2 Pathway. J Cancer. 2019;10:1375.
Tamburri S, Lavarone E, Fernández-Pérez D, Conway E, Zanotti M, Manganaro D, et al. Histone H2AK119 Mono-Ubiquitination Is Essential for Polycomb-Mediated Transcriptional Repression. Mol Cell. 2020;77:840–56.
Cohen I, Zhao D, Menon G, Nakayama M, Koseki H, Zheng D, et al. PRC1 preserves epidermal tissue integrity independently of PRC2. Genes Dev. 2019;33:55–60.
Nascimento-Filho CHV, Silveira EJD, Goloni-Bertollo EM, De Souza LB, Squarize CH, Castilho RM. Skin wound healing triggers epigenetic modifications of histone H4. J Transl Med. 2020;18:138.
Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37:391–400.
Qiang L, Sample A, Liu H, Wu X, He YY. Epidermal SIRT1 regulates inflammation, cell migration, and wound healing. Sci Rep.7:1–10.
Zhu H, Wang G, Qian J. Transcription factors as readers and effectors of DNA methylation. Nat Rev Genet. 2016;17:551–65.
Neri F, Rapelli S, Krepelova A, Incarnato D, Parlato C, Basile G, et al. Intragenic DNA methylation prevents spurious transcription initiation. Nature 2017;543:72–7.
Cedar H, Bergman Y. Epigenetics of haematopoietic cell development. Nat Rev Immunol. 2011;11:478–88.
Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998;19:219–20.
Moore LD, Le T, Fan G. DNA Methylation and Its Basic Function. Neuropsychopharmacology 2012;38:23–38.
Lauria A, Meng G, Proserpio V, Rapelli S, Maldotti M, Polignano IL, et al. DNMT3B supports meso-endoderm differentiation from mouse embryonic stem cells. Nat Commun. 2023;14:1–18.
Sen GL, Reuter JA, Webster DE, Zhu L, Khavari PA. DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature 2010;463:563–7.
Zhang J, Yang C, Wu C, Cui W, Wang L. DNA Methyltransferases in Cancer: Biology, Paradox, Aberrations, and Targeted Therapy. Cancers (Basel). 2020;12:1–22.
Etoh T, Kanai Y, Ushijima S, Nakagawa T, Nakanishi Y, Sasako M, et al. Increased DNA methyltransferase 1 (DNMT1) protein expression correlates significantly with poorer tumor differentiation and frequent DNA hypermethylation of multiple CpG islands in gastric cancers. Am J Pathol. 2004;164:689–99.
Luo G, Jing X, Yang S, Peng D, Dong J, Li L, et al. DNA Methylation Regulates Corneal Epithelial Wound Healing by Targeting miR-200a and CDKN2B. Invest Ophthalmol Vis Sci. 2019;60:650–60.
Yan J, Tie G, Wang S, Tutto A, Demarco N, Khair L, et al. Diabetes impairs wound healing by Dnmt1-dependent dysregulation of hematopoietic stem cells differentiation towards macrophages. Nat Commun. 2018;9:33.
Zhao C, Yang Q, Tang R, Li W, Wang J, Yang F. et al. DNA methyltransferase 1 deficiency improves macrophage motility and wound healing by ameliorating cholesterol accumulation. npj Regen Med. 2023;8:1–17.
Zhao J, Yang S, Shu B, Chen L, Yang R, Xu Y, et al. Transient High Glucose Causes Persistent Vascular Dysfunction and Delayed Wound Healing by the DNMT1-Mediated Ang-1/NF-κB Pathway. J Investig Dermatol. 2021;141:1573–84.
Bhatt T, Dey R, Hegde A, Ketkar AA, Pulianmackal AJ, Deb AP, et al. Initiation of wound healing is regulated by the convergence of mechanical and epigenetic cues. PLoS Biol. 2022;20:e3001777.
Leonard S, Pereira M, Fox R, Gordon N, Yap J, Kehoe S, et al. Over-expression of DNMT3A predicts the risk of recurrent vulvar squamous cell carcinomas. Gynecol Oncol. 2016;143:414–20.
Rinaldi L, Avgustinova A, Martín M, Datta D, Solanas G, Neus P, et al. Loss of Dnmt3a and Dnmt3b does not affect epidermal homeostasis but promotes squamous transformation through PPAR-γ. Elife 2017;6:e21697.
Yan F, Shen N, Pang J, Xie D, Deng B, Molina JR, et al. Restoration of miR-101 suppresses lung tumorigenesis through inhibition of DNMT3a-dependent DNA methylation. Cell Death Dis. 2014;5:e1413.
Gao Q, Steine EJ, Barrasa MI, Hockemeyer D, Pawlak M, Fu D, et al. Deletion of the de novo DNA methyltransferase Dnmt3a promotes lung tumor progression. Proc Natl Acad Sci USA. 2011;108:18061–6.
Rasmussen KD, Helin K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 2016;30:733.
Huang R, Wang Y, Ge H, Wang D, Wang Y, Zhang W, et al. Restoration of TET2 deficiency inhibits tumor growth in head neck squamous cell carcinoma. Ann Transl Med. 2020;8:329.
Zhou L, Ren M, Zeng T, Wang W, Wang X, Hu M, et al. TET2-interacting long noncoding RNA promotes active DNA demethylation of the MMP-9 promoter in diabetic wound healing. Cell Death Dis. 2019;10:1–13.
Wilson ER, Helton NM, Heath SE, Fulton RS, Payton JE, Welch JS, et al. Focal disruption of DNA methylation dynamics at enhancers in IDH-mutant AML cells. Leukemia 2021;36:935–45.
Hombach S, Kretz M. Non-coding RNAs: Classification, Biology and Functioning. Adv Exp Med Biol. 2016;937:3–17.
Luan A, Hu MS, Leavitt T, Brett EA, Wang KC, Longaker MT, et al. Noncoding RNAs in Wound Healing: A New and Vast Frontier. Adv Wound Care (N. Rochelle). 2018;7:19–27.
Li D, Niu G, Landén NX. Beyond the Code: Noncoding RNAs in Skin Wound Healing. 2022.
Sundaram GM, Quah S, Sampath P. Cancer: the dark side of wound healing. FEBS J 2018;285:4516–34.
Sun Y, Ma L. New Insights into Long Non-Coding RNA MALAT1 in Cancer and Metastasis. Cancers (Basel). 2019;11:216.
Liang ZH, Pan YC, Lin SS, Qiu ZY, Zhang Z. LncRNA MALAT1 promotes wound healing via regulating miR-141-3p/ZNF217 axis. Regen Ther. 2020;15:202.
Yang J, Qi M, Fei X, Wang X, Wang K. LncRNA H19: A novel oncogene in multiple cancers. Int J Biol Sci. 2021;17:3188.
Yu P, Guo J, Li J, Shi X, Xu N, Jiang Y, et al. lncRNA-H19 in Fibroblasts Promotes Wound Healing in Diabetes. Diabetes 2022;71:1562–78.
Tamagawa S, Beder LB, Hotomi M, Gunduz M, Yata K, Grenman R, et al. Role of miR-200c/miR-141 in the regulation of epithelial-mesenchymal transition and migration in head and neck squamous cell carcinoma. Int J Mol Med. 2014;33:879–86.
Sulaiman SA, Ab Mutalib NS, Jamal R. miR-200c Regulation of Metastases in Ovarian Cancer: Potential Role in Epithelial and Mesenchymal Transition. Front Pharm. 2016;7:271.
Aunin E, Broadley D, Ahmed MI, Mardaryev AN, Botchkareva NV. Exploring a Role for Regulatory miRNAs In Wound Healing during Ageing: Involvement of miR-200c in wound repair. Sci Rep. 2017;7:1–10.
Stojadinovic O, Ramirez H, Pastar I, Gordon KA, Stone R, Choudhary S, et al. MiR-21 and miR-205 are induced in invasive cutaneous squamous cell carcinomas. Arch Dermatol Res. 2017;309:133–9.
Arantes LMRB LausAC, Melendez ME, deCarvalho AC, Sorroche BP, De Marchi PRM. et al. MiR-21 as prognostic biomarker in head and neck squamous cell carcinoma patients undergoing an organ preservation protocol. Oncotarget. 2017;8:9911–21.
Hung PS, Tu HF, Kao SY, Yang CC, Liu CJ, Huang TY, et al. miR-31 is upregulated in oral premalignant epithelium and contributes to the immortalization of normal oral keratinocytes. Carcinogenesis 2014;35:1162–71.
Wang T, Feng Y, Sun H, Zhang L, Hao L, Shi C, et al. miR-21 regulates skin wound healing by targeting multiple aspects of the healing process. Am J Pathol. 2012;181:1911–20.
Li D, Li X, Wang A, Meisgen F, Pivarcsi A, Sonkoly E, et al. MicroRNA-31 Promotes Skin Wound Healing by Enhancing Keratinocyte Proliferation and Migration. J Invest Dermatol. 2015;135:1676–85.
Wilkinson E, Cui YH, He YY. Roles of RNA Modifications in Diverse Cellular Functions. Front Cell Dev Biol. 2022;10:828683.
Luo G, Xu W, Chen X, Xu W, Yang S, Wang J, et al. The RNA m5C Methylase NSUN2 Modulates Corneal Epithelial Wound Healing. Invest Ophthalmol Vis Sci. 2023;64:5.
Chellamuthu A, Gray SG. The RNA Methyltransferase NSUN2 and Its Potential Roles in Cancer. Cells NLM (Medlin). 2020;9:1758.
Frye M, Watt FM. The RNA methyltransferase Misu (NSun2) mediates Myc-induced proliferation and is upregulated in tumors. Curr Biol. 2006;16:971–81.
Zhou J, Wei T, He Z. ADSCs enhance VEGFR3-mediated lymphangiogenesis via METTL3-mediated VEGF-C m6A modification to improve wound healing of diabetic foot ulcers. Mol Med. 2021;27:146.
Cai J, Yang F, Zhan H, Situ J, Li W, Mao Y, et al. RNA m6A methyltransferase METTL3 promotes the growth of prostate cancer by regulating hedgehog pathway. Onco Targets Ther. 2019;12:9143–52.
Cai X, Wang X, Cao C, Gao Y, Zhang S, Yang Z, et al. HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7g. Cancer Lett. 2018;415:11–9.
Chen H, Gao S, Liu W, Wong CC, Wu J, Wu J, et al. RNA N6-Methyladenosine Methyltransferase METTL3 Facilitates Colorectal Cancer by Activating the m6A-GLUT1-mTORC1 Axis and Is a Therapeutic Target. Gastroenterology 2021;160:1284–300.
Arango D, Sturgill D, Alhusaini N, Meier JL, Coller J, Oberdoerffer S, et al. Acetylation of Cytidine in mRNA Promotes Translation Efficiency. Cell 2018;175:1872–86.
Xie L, Zhong X, Cao W, Liu J, Zu X, Chen L. Mechanisms of NAT10 as ac4C writer in diseases. Mol Ther Nucleic Acids. 2023;32:359–68.
Liao L, He Y, Li SJ, Yu XM, Liu ZC, Liang YY, et al. Lysine 2-hydroxyisobutyrylation of NAT10 promotes cancer metastasis in an ac4C-dependent manner. Cell Res. 2023;33:355–71.
Wang B, Zhang J, Li G, Xu C, Yang L, Zhang J, et al. N-acetyltransferase 10 promotes cutaneous wound repair via the NF-κB-IL-6 axis. Cell Death. Discovery 2023;9:1–10.
Giroux V, Rustgi AK. Metaplasia: tissue injury adaptation and a precursor to the dysplasia-cancer sequence. Nat Rev Cancer. 2017;17:594–604.
Fine JD, Johnson LB, Weiner M, Li KP, Suchindran C. Epidermolysis bullosa and the risk of life-threatening cancers: the National EB Registry experience, 1986-2006. J Am Acad Dermatol. 2009;60:203–11.
Wong SY, Reiter JF. Wounding mobilizes hair follicle stem cells to form tumors. Proc Natl Acad Sci USA. 2011;108:4093–8.
Kasper M, Jaks V, Are A, Bergström Å, Schwäger A, Barker N, et al. Wounding enhances epidermal tumorigenesis by recruiting hair follicle keratinocytes. Proc Natl Acad Sci USA. 2011;108:4099–104.
Higa T, Okita Y, Matsumoto A, Nakayama S, Oka T, Sugahara O, et al. Spatiotemporal reprogramming of differentiated cells underlies regeneration and neoplasia in the intestinal epithelium. Nat Commun. 2022;13:1500.
Schütte K, Bornschein J, Malfertheiner P. Hepatocellular carcinoma-epidemiological trends and risk factors. Dig Dis. 2009;27:80–92.
Riordan JD, Feddersen CR, Tschida BR, Beckmann PJ, Keng VW, Linden MA, et al. Chronic liver injury alters driver mutation profiles in hepatocellular carcinoma. Hepatology 2018;67:924.
Divangahi M, Aaby P, Abdul Khader S, Barreiro LB, Bekkering S, Chavakis T, et al. Trained immunity, tolerance, priming and differentiation: distinct immunological processes. Nat Immunol. 2021;22:2–6.
Kinoshita T, Seki M. Epigenetic memory for stress response and adaptation in plants. Plant Cell Physiol. 2014;55,1859–63.
Tehrani SSH, Kogan A, Mikulski P, Jansen LET. Remembering foods and foes: emerging principles of transcriptional memory. Cell Death Differ. 2023;1–11.
Netea MG, Domínguez-Andrés J, Barreiro LB, Chavakis T, Divangahi M, Fuchs E, et al. Defining trained immunity and its role in health and disease. Nat Rev Immunol. 2020;20:375–88.
Naik S, Larsen SB, Gomez NC, Alaverdyan K, Sendoel A, Yuan S, et al. Inflamm Mem sensitizes Ski Epithel stem cells tissue damage. 2017;550:475–80.
Larsen SB, Cowley CJ, Sajjath SM, Barrows D, Yang Y, Carroll TS, et al. Establishment, maintenance, and recall of inflammatory memory. Cell Stem Cell. 2021;28:1758–74.
Gonzales KAU, Polak L, Matos I, Tierney MT, Gola A, Wong E, et al. Stem cells expand potency and alter tissue fitness by accumulating diverse epigenetic memories. Science 2021;374:eabh2444.
Ordovas-Montanes J, Dwyer DF, Nyquist SK, Buchheit KM, Vukovic M, Deb C, et al. Allergic inflammatory memory in human respiratory epithelial progenitor cells. Nature 2018;560:649–54.
Lim AI, McFadden T, Link VM, Han SJ, Karlsson RM, Stacy A, et al. Prenatal maternal infection promotes tissue-specific immunity and inflammation in offspring. Science 2021;373:eabf3002.
Levra Levron C, Donati G. Multiplicity of stem cell memories of inflammation and tissue repair in epithelia. Trends Cell Biol. 2023;34:3–6.
Proserpio V, Oliviero S, Donati G. Locked and Loaded: Inflammation Training Prepares Skin Epithelial Stem Cells for Trauma. Cell Stem Cell. 2017;21:715–7.
Halper-Stromberg A, Jabri B. Maladaptive consequences of inflammatory events shape individual immune identity. Nat Immunol. 2022;23:1675–86.
Kadur Lakshminarasimha Murthy P, Xi R, Arguijo D, Everitt JI, Kocak DD, Kobayashi Y, et al. Epigenetic basis of oncogenic-Kras-mediated epithelial-cellular proliferation and plasticity. Dev Cell. 2022;57:310–28.
Mathilde Latil A, Nassar D, Beck B, Declercq W, Yi R, Blanpain C, et al. Cell-type-specific chromatin states differentially prime squamous cell carcinoma tumor-initiating cells for epithelial to mesenchymal transition. Cell Stem Cell. 2017;20:191–204.
Lochhead P, Chan AT, Nishihara R, Fuchs CS, Beck AH, Giovannucci E, et al. Etiologic field effect: Reappraisal of the field effect concept in cancer predisposition and progression. Mod Pathol. 2015;28:14–29.
Solé‐Boldo L, Raddatz G, Gutekunst J, Gilliam O, Bormann F, Liberio MS, et al. Differentiation-related epigenomic changes define clinically distinct keratinocyte cancer subclasses. Mol Syst Biol. 2022;18:e11073.
Slaughter DP, Southwick HW, Smejkal W. “Field cancerization” in oral stratified squamous epithelium. Clin Implic multicentric Orig Cancer. 1953;6:963–8.
Braakhuis BJM, Tabor MP, Kummer JA, Leemans CR, Brakenhoff RH. A Genetic Explanation of Slaughter’s Concept of Field Cancerization: Evidence and Clinical Implications 1. Cancer Res.2023;63:1727–30.
Alcolea MP, Greulich P, Wabik A, Frede J, Simons BD, Jones PH. Differentiation imbalance in single oesophageal progenitor cells causes clonal immortalization and field change. Nat Cell Biol. 2014;16:612–9.
Braakhuis BJM, Leemans CR, Brakenhoff RH. A genetic progression model of oral cancer: Current evidence and clinical implications. J Oral Pathol Med Blackwell Publ Ltd. 2004;33:317–22.
Sinjab A, Han G, Wang L, Kadara H. Field carcinogenesis in cancer evolution: What the cell is going on? Cancer Res. 2020;80:4888–91.
Katsurano M, Niwa T, Yasui Y, Shigematsu Y, Yamashita S, Takeshima H, et al. Early-stage formation of an epigenetic field defect in a mouse colitis model, and non-essential roles of T-and B-cells in DNA methylation induction. Oncogene 2012;31:342–51.
Nishiyama N, Arai E, Chihara Y, Fujimoto H, Hosoda F, Shibata T, et al. Genome-wide DNA methylation profiles in urothelial carcinomas and urothelia at the precancerous stage. Cancer Sci. 2010;101:231–40.
Baba Y, Ishimoto T, Kurashige J, Iwatsuki M, Sakamoto Y, Yoshida N, et al. Epigenetic field cancerization in gastrointestinal cancers. Cancer Lett. 2016;375:360–6.
Makabe T, Arai E, Hirano T, Ito N, Fukamachi Y, Takahashi Y, et al. Genome-wide DNA methylation profile of early-onset endometrial cancer: Its correlation with genetic aberrations and comparison with late-onset endometrial cancer. Carcinogenesis 2019;40:611–23.
Bernstein C. Epigenetic field defects in progression to cancer. World J Gastrointest Oncol. 2013;5:43.
Ramachandran K, Singal R. DNA methylation and field cancerization. Epigenomics 2012;4:243–5.
Španko M, Strnadová K, Pavlíček AJ, Szabo P, Kodet O, Valach J, et al. Il-6 in the ecosystem of head and neck cancer: Possible therapeutic perspectives. Int J Mol Sci. 2021;22:11027.
Wang J, He J, Zhu M, Han Y, Yang R, Liu H, et al. Cellular Heterogeneity and Plasticity of Skin Epithelial Cells in Wound Healing and Tumorigenesis. Stem Cell Rev Rep. 2022;18:1912.
Whitson RJ, Oro AE. Soil primes the seed: epigenetic landscape drives tumor behavior. Cell Stem Cell. 2017;20:149–50.
Lyko F. Distal memory in wound healing and cancer. Nat Cell Biol. 2023;25:631–2.
Donati G. The niche in single-cell technologies. Immunol Cell Biol. 2016;94:250–5.
Baysoy A, Bai Z, Satija R, Fan R. The technological landscape and applications of single-cell multi-omics. Nat Rev Mol Cell Biol. 2023;2023:1–19.
Toninelli M, Rossetti G, Pagani M. Charting the tumor microenvironment with spatial profiling technologies. Trends Cancer. 2023;9:1085-96.
Foster DS, Jones RE, Ransom RC, Longaker MT, Norton JA. The evolving relationship of wound healing and tumor stroma. JCI Insight. 2018;3:e99911.
Ghahramani A, Donati G, Luscombe NM, Watt FM. Epidermal Wnt signalling regulates transcriptome heterogeneity and proliferative fate in neighbouring cells. Genome Biol. 2018;19:1–14.
Armingol E, Officer A, Harismendy O, Lewis NE. Deciphering cell–cell interactions and communication from gene expression. Nat Rev Genet. 2020;22:71–88.
Van Hove L, Lecomte K, Roels J, Vandamme N, Vikkula H, Hoorens I, et al. Fibrotic enzymes modulate wound-induced skin tumorigenesis. EMBO Rep. 2021;22:e51573.
Hoste E, Arwert EN, Lal R, South AP, Salas-Alanis JC, Murrell DF, et al. Innate sensing of microbial products promotes wound-induced skin cancer. Nat Commun. 2015;6:5932.
Hu B, Castillo E, Harewood L, Ostano P, Reymond A, Dummer R, et al. Multifocal epithelial tumors and field cancerization from loss of mesenchymal CSL signaling. Cell 2012;149:1207–20.
Acknowledgements
Apologies to authors whose work could not be included due to space constraints. The Donati laboratory is supported by the Chan Zuckerberg Initiative (Single-Cell Analysis of Inflammation, Id. DAF2020-217532, https://doi.org/10.37921/173068fmftmj) and by AIRC, Associazione Italiana per la Ricerca sul Cancro (MFAG 2018 - Id. 21640).
Author information
Authors and Affiliations
Contributions
CLL and GD conceptualized the idea of the review. CLL, LE, and GD prepared the initial drafts of the manuscript. CD, GP, and VP contributed to the writing and manuscript improvement. CLL, LE, and GD contributed to the creation of figures and tables. All authors reviewed the manuscript and approved to the final version of this manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Levra Levron, C., Elettrico, L., Duval, C. et al. Bridging tissue repair and epithelial carcinogenesis: epigenetic memory and field cancerization. Cell Death Differ (2024). https://doi.org/10.1038/s41418-023-01254-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41418-023-01254-6