Abstract
Background
This study aims to develop and validate an artificial intelligence (AI)-aided Prostate Imaging Reporting and Data System (PI-RADSAI) for prostate cancer (PCa) diagnosis based on MRI.
Methods
The deidentified MRI data of 1540 biopsy-naïve patients were collected from four centres. PI-RADSAI is a two-stage, human-in-the-loop AI capable of emulating the diagnostic acumen of subspecialists for PCa on MRI. The first stage uses a UNet-Seg model to detect and segment biopsy-candidate prostate lesions, whereas the second stage leverages UNet-Seg segmentation is trained specifically with subspecialist’ knowledge-guided 3D-Resnet to achieve an automatic AI-aided diagnosis for PCa.
Results
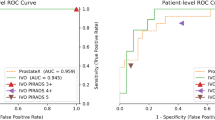
In the independent test set, UNet-Seg identified 87.2% (628/720) of target lesions, with a Dice score of 44.9% (range, 22.8–60.2%) in segmenting lesion contours. In the ablation experiment, the model trained with the data from three centres was superior (kappa coefficient, 0.716 vs. 0.531) to that trained with single-centre data. In the internal and external tests, the triple-centre PI-RADSAI model achieved an overall agreement of 58.4% (188/322) and 60.1% (92/153) with a referential subspecialist in scoring target lesions; when one-point margin of error was permissible, the agreement rose to 91.3% (294/322) and 97.3% (149/153), respectively. In the paired test, PI-RADSAI outperformed 5/11 (45.5%) and matched the performance of 3/11 (27.3%) general radiologists in achieving a clinically significant PCa diagnosis (area under the curve, internal test, 0.801 vs. 0.770, p < 0.01; external test, 0.833 vs. 0.867, p = 0.309).
Conclusions
Our closed-loop PI-RADSAI outperforms or matches the performance of more than 70% of general readers in the MRI assessment of PCa. This system might provide an alternative to radiologists and offer diagnostic benefits to clinical practice, especially where subspecialist expertise is unavailable.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
$259.00 per year
only $10.79 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Deidentified blinded raw data used to conduct the retrospective and prospective analyses are available upon request to ZY. The source code of the model is archived on GitHub (https://github.com/yuruiqi/PI-RADS_Classification). Requests for the raw images and associated DICOM data used to train and evaluate the model can be directed to Y-DZ, but will only be granted after specific IRB approvals and bespoke data agreement is established between the hospital health network and the requesting party.
References
Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7–33.
Mottet N, van den Bergh RCN, Briers E, Van den Broeck T, Cumberbatch MG, De Santis M, et al. EAU-EANM-ESTRO-ESUR-SIOG Guidelines on Prostate Cancer-2020 Update. Part 1: Screening, Diagnosis, and Local Treatment with Curative Intent. Eur Urol. 2021;79:243–62. https://doi.org/10.1016/j.eururo.2020.09.042.
Lin K, Lipsitz R, Miller T, Janakiraman S, Force USPST. Benefits and harms of prostate-specific antigen screening for prostate cancer: an evidence update for the U.S. Preventive Services Task Force. Ann Intern Med. 2008;149:192–9. https://doi.org/10.7326/0003-4819-149-3-200808050-00009.
Ilic D, Neuberger MM, Djulbegovic M, Dahm P. Screening for prostate cancer. Cochr Database Syst Rev. 2013. https://doi.org/10.1002/14651858.CD004720.pub3.
Schroder FH, Hugosson J, Roobol MJ, Tammela TL, Ciatto S, Nelen V, et al. Screening and prostate-cancer mortality in a randomized European study. N Engl J Med. 2009;360:1320–8. https://doi.org/10.1056/NEJMoa0810084.
Zhu X, Albertsen PC, Andriole GL, Roobol MJ, Schroder FH, Vickers AJ. Risk-based prostate cancer screening. Eur Urol. 2012;61:652–61. https://doi.org/10.1016/j.eururo.2011.11.029.
Wang R, Wang J, Gao G, Hu J, Jiang Y, Zhao Z, et al. Prebiopsy mp-MRI can help to improve the predictive performance in prostate cancer: a prospective study in 1,478 consecutive patients. Clin Cancer Res. 2017;23:3692–9. https://doi.org/10.1158/1078-0432.CCR-16-2884.
Ahmed HU, El-Shater Bosaily A, Brown LC, Gabe R, Kaplan R, Parmar MK, et al. Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMIS): a paired validating confirmatory study. Lancet. 2017;389:815–22. https://doi.org/10.1016/S0140-6736(16)32401-1.
Park SY, Jung DC, Oh YT, Cho NH, Choi YD, Rha KH, et al. Prostate cancer: PI-RADS Version 2 helps preoperatively predict clinically significant cancers. Radiology. 2016;280:108–16. https://doi.org/10.1148/radiol.16151133.
Ahmed HU, Kirkham A, Arya M, Illing R, Freeman A, Allen C, et al. Is it time to consider a role for MRI before prostate biopsy? Nat Rev Clin Oncol. 2009;6:197–206. https://doi.org/10.1038/nrclinonc.2009.18.
Gupta RT, Mehta KA, Turkbey B, Verma S. PI-RADS: past, present, and future. J Magn Reson Imaging. 2020;52:33–53. https://doi.org/10.1002/jmri.26896.
Kasivisvanathan V, Rannikko AS, Borghi M, Panebianco V, Mynderse LA, Vaarala MH, et al. MRI-Targeted or standard biopsy for prostate-cancer diagnosis. N Engl J Med. 2018;378:1767–77. https://doi.org/10.1056/NEJMoa1801993.
Oberlin DT, Casalino DD, Miller FH, Meeks JJ. Dramatic increase in the utilization of multiparametric magnetic resonance imaging for detection and management of prostate cancer. Abdom Radiol (N. Y). 2017;42:1255–8. https://doi.org/10.1007/s00261-016-0975-5.
Girometti R, Giannarini G, Greco F, Isola M, Cereser L, Como G, et al. Interreader agreement of PI-RADS v. 2 in assessing prostate cancer with multiparametric MRI: a study using whole-mount histology as the standard of reference. J Magn Reson Imaging. 2019;49:546–55. https://doi.org/10.1002/jmri.26220.
Thai JN, Narayanan HA, George AK, Siddiqui MM, Shah P, Mertan FV, et al. Validation of PI-RADS version 2 in transition zone lesions for the detection of prostate cancer. Radiology. 2018;288:485–91. https://doi.org/10.1148/radiol.2018170425.
Shankar PR, Kaza RK, Al-Hawary MM, Masch WR, Curci NE, Mendiratta-Lala M, et al. Impact of clinical history on maximum PI-RADS Version 2 Score: a six-reader 120-case sham history retrospective evaluation. Radiology. 2018;288:158–63. https://doi.org/10.1148/radiol.2018172619.
Padhani AR, Turkbey B. Detecting prostate cancer with deep learning for MRI: a small step forward. Radiology. 2019;293:618–9. https://doi.org/10.1148/radiol.2019192012.
Rosenkrantz AB, Ginocchio LA, Cornfeld D, Froemming AT, Gupta RT, Turkbey B, et al. Interobserver reproducibility of the PI-RADS Version 2 Lexicon: a multicenter study of six experienced prostate radiologists. Radiology. 2016;280:793–804. https://doi.org/10.1148/radiol.2016152542.
Smith CP, Harmon SA, Barrett T, Bittencourt LK, Law YM, Shebel H, et al. Intra- and interreader reproducibility of PI-RADSv2: a multireader study. J Magn Reson Imaging. 2019;49:1694–703. https://doi.org/10.1002/jmri.26555.
Brembilla G, Dell’Oglio P, Stabile A, Damascelli A, Brunetti L, Ravelli S, et al. Interreader variability in prostate MRI reporting using Prostate Imaging Reporting and Data System version 2.1. Eur Radiol. 2020;30:3383–92. https://doi.org/10.1007/s00330-019-06654-2.
Wildeboer RR, van Sloun RJG, Wijkstra H, Mischi M. Artificial intelligence in multiparametric prostate cancer imaging with focus on deep-learning methods. Computer Methods Prog Biomed. 2020;189:105316 https://doi.org/10.1016/j.cmpb.2020.105316.
Roethke MC, Kuru TH, Mueller-Wolf MB, Agterhuis E, Edler C, Hohenfellner M, et al. Evaluation of an automated analysis tool for prostate cancer prediction using multiparametric magnetic resonance imaging. PLoS ONE. 2016;11:e0159803 https://doi.org/10.1371/journal.pone.0159803.
Litjens G, Debats O, Barentsz J, Karssemeijer N, Huisman H. Computer-aided detection of prostate cancer in MRI. IEEE Trans Med Imaging. 2014;33:1083–92. https://doi.org/10.1109/TMI.2014.2303821.
Schelb P, Kohl S, Radtke JP, Wiesenfarth M, Kickingereder P, Bickelhaupt S, et al. Classification of Cancer at Prostate MRI: Deep Learning versus Clinical PI-RADS Assessment. Radiology. 2019;293:607–17. https://doi.org/10.1148/radiol.2019190938.
Sanford T, Harmon SA, Turkbey EB, Kesani D, Tuncer S, Madariaga M, et al. Deep-learning-based artificial intelligence for PI-RADS classification to assist multiparametric prostate MRI interpretation: a development study. J Magn Reson Imaging. 2020. https://doi.org/10.1002/jmri.27204.
Saha A, Hosseinzadeh M, Huisman H. End-to-end prostate cancer detection in bpMRI via 3D CNNs: Effects of attention mechanisms, clinical priori and decoupled false positive reduction. Med Image Anal. 2021;73:102155 https://doi.org/10.1016/j.media.2021.102155.
Winkel DJ, Tong A, Lou B, Kamen A, Comaniciu D, Disselhorst JA, et al. A novel deep learning based computer-aided diagnosis system improves the accuracy and efficiency of radiologists in reading biparametric magnetic resonance images of the prostate: results of a multireader, multicase study. Investig Radiol. 2021;56:605–13. https://doi.org/10.1097/RLI.0000000000000780.
Litjens G, Debats O, Barentsz J, Karssemeijer N, Huisman H. Computer-aided detection of prostate cancer in MRI. IEEE Trans Med Imaging. 2014;33:1083–92. https://doi.org/10.1109/TMI.2014.2303821.
Adams LC, Makowski MR, Engel G, Rattunde M, Busch F, Asbach P, et al. Prostate158—an expert-annotated 3T MRI dataset and algorithm for prostate cancer detection. Comput Biol Med. 2022;148:105817 https://doi.org/10.1016/j.compbiomed.2022.105817.
Hansen N, Patruno G, Wadhwa K, Gaziev G, Miano R, Barrett T, et al. Magnetic resonance and ultrasound image fusion supported transperineal prostate biopsy using the ginsburg protocol: technique, learning points, and biopsy results. Eur Urol. 2016;70:332–40. https://doi.org/10.1016/j.eururo.2016.02.064.
Epstein JI, Egevad L, Amin MB, Delahunt B, Srigley JR, Humphrey PA, et al. The 2014 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma: Definition of Grading Patterns and Proposal for a New Grading System. Am J surgical Pathol. 2016;40:244–52. https://doi.org/10.1097/PAS.0000000000000530.
Jianlin C, Zheng W, Pollastri G. A neural network approach to ordinal regression. In: 2008 IEEE International Joint Conference on Neural Networks. IEEE World Congress on Computational Intelligence; 2008.
Vente CD, Vos P, Hosseinzadeh M, Pluim J, Veta M. Deep learning regression for prostate cancer detection and grading in bi-parametric MRI. IEEE Trans Biomed Eng. 2021;68:374–83. https://doi.org/10.1109/TBME.2020.2993528.
Wu B, Sun X, Hu L, Wang Y. Learning with unsure data for medical image diagnosis. In: Proc. IEEE/CVF International Conference on Computer Vision. 2019, pp. 10589–98. https://doi.org/10.1109/ICCV.2019.01069.
Vasey B, Nagendran M, Campbell B, Clifton DA, Collins GS, Denaxas S, et al. Reporting guideline for the early-stage clinical evaluation of decision support systems driven by artificial intelligence: DECIDE-AI. Nat Med. 2022;28:924–33. https://doi.org/10.1038/s41591-022-01772-9.
Kvamme H, Borgan Ø. Continuous and discrete-time survival prediction with neural networks. Lifetime Data Anal. 2021;27:710–36. https://doi.org/10.1007/s10985-021-09532-6.
Futterer JJ, Briganti A, De Visschere P, Emberton M, Giannarini G, Kirkham A, et al. Can clinically significant prostate cancer be detected with multiparametric magnetic resonance imaging? A systematic review of the literature. Eur Urol. 2015;68:1045–53. https://doi.org/10.1016/j.eururo.2015.01.013.
Thompson JE, Moses D, Shnier R, Brenner P, Delprado W, Ponsky L, et al. Multiparametric magnetic resonance imaging guided diagnostic biopsy detects significant prostate cancer and could reduce unnecessary biopsies and over detection: a prospective study. J Urol. 2014;192:67–74. https://doi.org/10.1016/j.juro.2014.01.014.
Acknowledgements
We thank all those who helped us during the writing of this research. We also thank the department of Ultrasound, Urology and Pathology of the hospitals for their valuable help and feedback.
Funding
This work is supported by the National Natural Science Foundation of China (contract grant number: 61731009, GY; 82272082, Y-DZ).
Author information
Authors and Affiliations
Contributions
Study conception: YH, YS, GY and Y-DZ. Data collection: KJ, JB, YH, C-HH and Y-DZ. Data analysis and interpretation: RY, KJ, JB, YH, YY, DW, YS, C-HH, GY and Y-DZ. Technical support: RY, YY, DW, YS, GY and Y-DZ. Administrative support: C-HH, GY and Y-DZ. Manuscript drafting: RY, KJ, GY and Y-DZ. All authors read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Ethics approval for the use of data from centre 1 and centre 2 was granted by the hospital institutional review board (grant no., 2019-SR-396), and informed patient consent was waived.
Consent to publish
Not applicable.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yu, R., Jiang, Kw., Bao, J. et al. PI-RADSAI: introducing a new human-in-the-loop AI model for prostate cancer diagnosis based on MRI. Br J Cancer 128, 1019–1029 (2023). https://doi.org/10.1038/s41416-022-02137-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-022-02137-2
This article is cited by
-
Improved prostate cancer diagnosis using a modified ResNet50-based deep learning architecture
BMC Medical Informatics and Decision Making (2024)
-
Comment on “Pathological complete response, category change, and prognostic significance of HER2-low breast cancer receiving neoadjuvant treatment: a multicenter analysis of 2489 cases”
British Journal of Cancer (2024)
-
What benefit can be obtained from magnetic resonance imaging diagnosis with artificial intelligence in prostate cancer compared with clinical assessments?
Military Medical Research (2023)