Abstract
Background
Prostate cancer is a clinically and molecularly heterogeneous disease, with highest incidence and mortality among men of African ancestry. To date, prostate cancer patient-derived xenograft (PCPDX) models to study this disease have been difficult to establish because of limited specimen availability and poor uptake rates in immunodeficient mice. Ancestrally diverse PCPDXs are even more rare, and only six PCPDXs from self-identified African American patients from one institution were recently made available.
Methods
In the present study, we established a PCPDX from prostate cancer tissue from a patient of estimated 90% West African ancestry with metastatic castration resistant disease, and characterized this model’s pathology, karyotype, hotspot mutations, copy number, gene fusions, gene expression, growth rate in normal and castrated mice, therapeutic response, and experimental metastasis.
Results
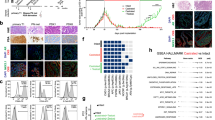
This PCPDX has a mutation in TP53 and loss of PTEN and RB1. We have documented a 100% take rate in mice after thawing the PCPDX tumor from frozen stock. The PCPDX is castrate- and docetaxel-resistant and cisplatin-sensitive, and has gene expression patterns associated with such drug responses. After tail vein injection, the PCPDX tumor cells can colonize the lungs of mice.
Conclusion
This PCPDX, along with others that are established and characterized, will be useful pre-clinically for studying the heterogeneity of prostate cancer biology and testing new therapeutics in models expected to be reflective of the clinical setting.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 4 print issues and online access
$259.00 per year
only $64.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All RNA-sequencing data are available from GEO (GSE146402).
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. Cancer J Clin. 2019;69:7–34.
Cancer Genome Atlas Research Network. The molecular taxonomy of primary prostate cancer. Cell. 2015;163:1011–25.
Irshad S, Bansal M, Castillo-Martin M, Zheng T, Aytes A, Wenske S, et al. A molecular signature predictive of indolent prostate cancer [published correction appears in Sci Transl Med. 2013 Sep 18;5(203):203er9]. Sci Transl Med. 2013;5:202ra122.
Robbins AS, Whittemore AS, Thom DH. Differences in socioeconomic status and survival among white and black men with prostate cancer. Am J Epidemiol. 2000;151:409–16.
Pietro GD, Chornokur G, Kumar NB, Davis C, Park JY. Racial differences in the diagnosis and treatment of prostate cancer. Int Neurourol J. 2016;20:S112–S119. https://doi.org/10.5213/inj.1632722.361
Polite BN, Adams-Campbell LL, Brawley OW, Bickell N, Carethers JM, Flowers CR, et al. Charting the future of cancer health disparities research: a position statement from the American Association for Cancer Research, the American Cancer Society, the American Society of Clinical Oncology, and the National Cancer Institute. CA Cancer J Clin. 2017;67:353–61.
Navone NM, van Weerden WM, Vessella RL, Williams ED, Wang Y, Isaacs JT, et al. Movember gap1 pdx project: an international collection of serially transplantable prostate cancer patient-derived xenograft (pdx) models. Prostate. 2018;78:1262–82.
Palanisamy N, Yang J, Shepherd PDA, Li-Ning-Tapia EM, Labanca E, Manyam GC, et al. The MD Anderson prostate cancer patient-derived xenograft series (MDA PCa PDX) captures the molecular landscape of prostate cancer and facilitates marker-driven therapy development. Clin Cancer Res. 2020. https://doi.org/10.1158/1078-0432.ccr-20-0479.
Bluemn EG, Coleman IM, Lucas JM, Coleman RT, Hernandez-Lopez S, Tharakan R, et al. Androgen receptor pathway-independent prostate cancer is sustained through FGF signaling. Cancer Cell. 2017;32:474–89. e6.
Gao H, Korn JM, Ferretti S, Monahan JE, Wang Y, Singh M, et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat Med. 2015;21:1318–25. https://doi.org/10.1038/nm.3954.
Russell PJ, Russell P, Rudduck C, Tse BW, Williams ED, Raghavan D. Establishing prostate cancer patient derived xenografts: lessons learned from older studies. Prostate. 2015;75:628–36. https://doi.org/10.1002/pros.22946.
Zhao H, Nolley R, Chen Z, Peehl DM. Tissue slice grafts: an in vivo model of human prostate androgen signaling. Am J Pathol. 2010;177:229–39. https://doi.org/10.2353/ajpath.2010.090821.
Zhao H, Thong A, Nolley R, Reese SW, Santos J, Ingles A, et al. Patient-derived tissue slice grafts accurately depict response of high-risk primary prostate cancer to androgen deprivation therapy. J Transl Med. 2013;11:199. https://doi.org/10.1186/1479-5876-11-199.
Wang Y, Revelo MP, Sudilovsky D, Cao M, Chen WG, Goetz L, et al. Development and characterization of efficient xenograft models for benign and malignant human prostate tissue. Prostate. 2005;64:149–59.
Priolo C, Agostini M, Vena N, Ligon AH, Fiorentino M, Shin E, et al. Establishment and genomic characterization of mouse xenografts of human primary prostate tumors. Am J Pathol. 2010;176:1901–13.
Lin D, Wyatt AW, Xue H, Wang Y, Dong X, Haegart A. et al. High fidelity patient-derived xenografts for accelerating prostate cancer discovery and drug development. Cancer Res. 2014;74:1272–83. https://doi.org/10.1158/0008-5472.CAN-13-2921-T.
Goldstein AS, Drake JM, Burnes DL, Finley DS, Zhang H, Reiter RE, et al. Purification and direct transformation of epithelial progenitor cells from primary human prostate. Nat Protoc. 2011;6:656–67.
Uronis JM, Osada T, McCall S, Yang XY, Mantyh C, Morse MA, et al. Histological and molecular evaluation of patient-derived colorectal cancer explants. PLoS One. 2012;7:e38422.
Kim MK, Osada T, Barry WT, Yang XY, Freedman JA, Tsamis KA, et al. Characterization of an oxaliplatin sensitivity predictor in a preclinical murine model of colorectal cancer. Mol Cancer Ther. 2012;11:1500–9.
Nguyen HM, Vessella RL, Morrissey C, Brown LG, Coleman IM, Higano CS, et al. LuCaP prostate cancer patient-derived xenografts reflect the molecular heterogeneity of advanced disease an-d serve as models for evaluating cancer therapeutics. Prostate. 2017;77:654–71.
Lawrence MG, Obinata D, Sandhu S. Patient-derived models of abiraterone- and enzalutamide-resistant prostate cancer reveal sensitivity to ribosome-directed therapy. Eur Urol. 2018;74:562–72. https://doi.org/10.1016/j.eururo.2018.06.020.
Li Q, Deng Q, Chao HP, Liu X, Lu Y, Lin K, et al. Linking prostate cancer cell AR heterogeneity to distinct castration and enzalutamide responses. Nat Commun. 2018;9:3600.
Pienta KJ, Abate-Shen C, Agus DB, Attar RM, Chung LWK, Greenberg NM, et al. The current state of preclinical prostate cancer animal models. Prostate. 2008;68:629–39.
Wise JP Sr, Wise SS, Little JE. The cytotoxicity and genotoxicity of particulate and soluble hexavalent chromium in human lung cells. Mutat Res. 2002;517:221–9.
Al-Alem U, Rauscher G, Shah E, Batai K, Mahmoud A, Beisner E, et al. Association of genetic ancestry with breast cancer in ethnically diverse women from Chicago. PLoS ONE. 2014;9:e112916.
Gupta S, Li J, Kemeny G, Bitting RL, Beaver J, Somarelli JA, et al. Whole genomic copy number alterations in circulating tumor cells from men with abiraterone or enzalutamide-resistant metastatic castration-resistant prostate cancer. Clin Cancer Res. 2017;23:1346–57. https://doi.org/10.1158/1078-0432.CCR-16-1211.
Andrews S. FastQC a quality control tool for high throughput sequence data. 2014. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
Ewels P, Magnusson M, Lundin S, Kaller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–8.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski Z, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21.
Anders S, Pyl PT, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9.
Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106.
Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J R Stat Soc Ser B-Methodol. 1995;57:289–300.
Bass A, Storey J. qvalue: Q-value estimation for false discovery rate control. R (published bioinformatics package) package version 2.8.0, 2015. http://github.com/jdstorey/qvalue.
Haas B, Dobin A, Stransky N, Li B, Yang X, Tickle T, et al. STAR-fusion: fast and accurate fusion transcript detection from RNA-Seq. bioRxiv. 2017. https://doi.org/10.1101/120295.
Lagstad S. chimeraviz: visualization tools for gene fusions. R package version 1.10.0, 2019. https://www.bioconductor.org/packages/release/bioc/html/chimeraviz.html.
R Core Team. R: a language and environment for statistical computing. Vienna, Austria; R Core Team: 2019.
Gentlema R, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80.
Xie Y. Dynamic documents with R and knitr. 2nd ed. Boca Raton: CRC Press/Taylor and Francis; 2015. xxvii, 266.
Chang JT, Nevins JR. GATHER: a systems approach to interpreting genomic signatures. Bioinformatics. 2006;22:2926–33. https://doi.org/10.1093/bioinformatics/btl483.
Krämer A, Green J, Pollard J Jr, Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30:523–30.
Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–28.
Zhu Z, Chung YM, Sergeeva O, Kepe V, Berk M, Li J, et al. Loss of dihydrotestosterone-inactivation activity promotes prostate cancer castration resistance detectable by functional imagingJ. Biol Chem. 2018;293:17829.
Leinonen KA, Saramäki OR, Furusato B, Kimura T, Takahashi H, Egawa S, et al. Loss of PTEN is associated with aggressive behavior in ERG-positive prostate cancer. Cancer Epidemiol Biomark Prev. 2013;22:2333–44.
Kregel S, Kiriluk KJ, Rosen AM, Cai Y, Reyes EE, Otto KB, et al. Sox2 is an androgen receptor-repressed gene that promotes castration-resistant prostate cancer. PLoS ONE. 2013;8:e53701.
Mu P, Zhang Z, Benelli M, Karthaus W, Hoover E, Chen C, et al. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science. 2017;355:84–88. https://doi.org/10.1126/science.aah4307.
Lee M, Williams KA, Hu Y, Andreas J, Patel SJ, Zhang S, et al. GNL3 and SKA3 are novel prostate cancer metastasis susceptibility genes. Clin Exp Metastasis. 2015;32:769–82.
Magi-Galluzzi C, Tsusuki T, Elson P, Simmerman K, LaFargue C, Esgueva R, et al. TMPRSS2-ERG gene fusion prevalence and class are significantly different in prostate cancer of Caucasian, African-American and Japanese patients. Prostate. 2011;71:489–97.
Crea F, Quagliata L, Michael A, Liu H, Frumento P, Azad AA. et al. Integrated analysis of the prostate cancer small-nucleolar transcriptome reveals SNORA55 as a driver of prostate cancer progression. Mol Oncol. 2016;10:693–703. https://doi.org/10.1016/j.molonc.2015.12.010.
Rocha CRR, Silva MM, Quinet A, Cabral-Neto JB, Menck CFM. DNA repair pathways and cisplatin resistance: an intimate relationship. Clinics. 2018;73:e478s. https://doi.org/10.6061/clinics/2018/e478s.
Abida W, Cyrta J, Heller G, Prandi D, Armenia J, Coleman I. et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc Natl Acad Sci USA. 2019;116:11428–36. https://doi.org/10.1073/pnas.1902651116.
Xu L, Tang H, Wang K, Zheng Y, Feng J, Dong H, et al. Pharmacological inhibition of EZH2 combined with DNA-damaging agents interferes DNA damage response MM cells. Mol Med Rep. 2019;19:4249–55.
Zhan J, Wang P, Li S, Song J, He H, Wang Y. et al. HOXB13 networking with ABCG1/EZH2/Slug mediates metastasis and confers resistance to cisplatin in lung adenocarcinoma patients. Theranostics. 2019;9:2084–99. https://doi.org/10.7150/thno.29463. 76.
Qin X, Guo H, Wang X, Zhu X, Yan M, Wang X, et al. Exosomal miR-196a derived from cancer-associated fibroblasts confers cisplatin resistance in head and neck cancer through targeting CDKN1B and ING5. Genome Biol. 2019;20:12. https://doi.org/10.1186/s13059-018-1604-0.
Shen DW, Pouliot LM, Gillet JP, Ma W, Johnson AC, Hall MD, et al. The transcription factor GCF2 is an upstream repressor of the small GTPAse RhoA, regulating membrane protein trafficking, sensitivity to doxorubicin, and resistance to cisplatin. Mol Pharm. 2012;9:1822–33. https://doi.org/10.1021/mp300153z.
Street CA, Routhier AA, Spencer C, Perkins AL, Masterjohn K, Hackathorn A, et al. Pharmacological inhibition of Rho-kinase (ROCK) signaling enhances cisplatin resistance in neuroblastoma cells. Int J Oncol. 2010;37:1297–305. https://doi.org/10.3892/ijo_00000781.
Yu C, Wu G, Li R, Gao L, Yang F, Zhao Y, et al. NDRG2 acts as a negative regulator downstream of androgen receptor and inhibits the growth of androgen-dependent and castration-resistant prostate cancer. Cancer Biol Ther. 2015;16: 287–96. https://doi.org/10.1080/15384047.2014.1002348.
Elvenes J, Thomassen EI, Johnsen SS, Kaino K, Sjøttem E, Johansen T. Pax6 represses androgen receptor-mediated transactivation by inhibiting recruitment of the coactivator SPBP. PloS One. 2011;6:e24659. https://doi.org/10.1371/journal.pone.0024659.
Shang Z, Niu Y, Cai Q, Chen J, Tian J, Yeh S. et al. Human kallikrein 2 (KLK2) promotes prostate cancer cell growth via function as a modulator to promote the ARA70-enhanced androgen receptor transactivation. Tumor Biol. 2014;35:1881–90. https://doi.org/10.1007/s13277-013-1253-6.
Patel R, Brzezinska EA, Repiscak P, Ahmad I, Mui E, Gao M, et al. Activation of β-catenin cooperates with loss of Pten to drive AR-independent castration-resistant prostate cancer. Cancer Res. 2020; 576–90. https://doi.org/10.1158/0008-5472.CAN-19-1684.
Nesbitt H, Browne G, O’Donovan KM, Byrne NM, Worthington J, McKeown SR. et al. Nitric oxide up-regulates RUNX2 in LNCaP prostate tumours: implications for tumour growth in vitro and in vivo. J Cell Physiol. 2016;23:473–82. https://doi.org/10.1002/jcp.25093.
Yang Y, Bai Y, He Y, Zhao Y, Chen J, Ma L. et al. PTEN loss promotes intratumoral androgen synthesis and tumor microenvironment remodeling via aberrant activation of RUNX2 in castration-resistant prostate cancer. Clin Cancer Res. 2018;24:834–46. https://doi.org/10.1158/1078-0432.CCR-17-2006.
Ozaki T, Yu M, Yin D, Sun D, Zhu Y, Bu Y, et al. Impact of RUNX2 on drug-resistant human pancreatic cancer cells with p53 mutations. BMC Cancer. 2018;18:309. https://doi.org/10.1186/s12885-018-4217-9.
Yu L, Su YS, Zhao J, Wang H, Li W. Repression of NR4A1 by a chromatin modifier promotes docetaxel resistance in PC-3 human prostate cancer cells. FEBS Lett. 2013;587:2542–51.
Elkin M, Vlodavsky I. Tail vein assay of cancer metastasis. Curr Protoc Cell Biol. 2001. https://doi.org/10.1002/0471143030.cb1902s12.
Acknowledgements
We acknowledge the BioRepository & Precision Pathology Center (BRPC), a shared resource of the Duke University School of Medicine and Duke Cancer Institute, for providing access to the human biospecimens used under Institutional Review Board oversight in this work, the assistance of the Duke University Health System Clinical Molecular Diagnostics Laboratory, Duke Sequencing and Genomic Technologies Shared Resource, and the Duke Cancer Institute Bioinformatics Shared Resource, and Bonnie LaCroix, laboratory manager. Support: P30 Cancer Center Support Grant (P30 CA014236), NIH Basic Research in Cancer Health Disparities R01 Award R01CA220314 to SRP PI, JAF and DSH Co-I, DJG and KO Collaborator, WCF Pathologist.
Author information
Authors and Affiliations
Contributions
BMP: Conceptualization, Investigation, Formal Analysis, Writing, Visualization; WCF: Investigation, Formal Analysis, Writing, Visualization; TA: Formal Analysis, Writing, Visualization; JAS: Methodology, Writing; KEW: Methodology, Writing; SG: Investigation, Formal Analysis, Writing, Visualization; SW: Investigation, Formal Analysis, Writing, Visualization; JPW: Methodology, Writing; XQ: Formal Analysis, Writing, Visualization; DZ: Formal Analysis, Writing, Visualization; LX: Methodology; YL: Methodology; XC: Methodology; BAI: Conceptualization, Resources, Writing; SJM: Resources, Writing; JH: Conceptualization, Writing; RAK: Investigation, Formal Analysis, Writing, Visualization; KO: Conceptualization, Validation, Writing; SG: Conceptualization, Validation, Writing; AJA: Conceptualization, Writing; DJG: Conceptualization, Writing; SRP: Conceptualization, Writing, Supervision, Funding Acquisition; DSH: Conceptualization, Writing, Supervision, Project Administration, Funding Acquisition; JAF: Conceptualization, Writing, Supervision, Project Administration, Funding Acquisition.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Patierno, B.M., Foo, WC., Allen, T. et al. Characterization of a castrate-resistant prostate cancer xenograft derived from a patient of West African ancestry. Prostate Cancer Prostatic Dis 25, 513–523 (2022). https://doi.org/10.1038/s41391-021-00460-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41391-021-00460-y
This article is cited by
-
The future of patient-derived xenografts in prostate cancer research
Nature Reviews Urology (2023)