Abstract
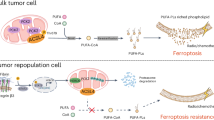
Most forms of chemotherapy for acute myeloid leukemia (AML) are often ineffective in eliminating leukemic stem cells (LSCs), as their underlying mechanisms remain unclear. Here, we have identified circFAM193B, which regulates the redox biology of LSCs and is associated with unfavorable outcomes in AML patients. In vitro and in vivo assays suggested that circFAM193B significantly inhibits LSCs chemotherapy resistance and AML progression. Knockdown circFAM193B enhances mitochondrial OXPHOS function and inhibits the accumulation of reactive oxygen species and lipid peroxidation mediated by chemotherapy, which protects AML cells from oxidative stress-induced cell death. Mechanistically, circFAM193B physically interacts with arginine methyltransferase PRMT6 catalytic domain and enhances the transcription efficiency of key lipid peroxidation factor ALOX15 by decreasing H3R2me2a modification. In summary, we have identified circFAM193B was downregulated in LSCs to promote the survival of LSC by modulating energy metabolism and the redox balance in the postchemotherapy persistence of LSC. Our studies provide a conceptual advance and biological insights regarding the drug resistance of LSCs via circRNA mediated PRMT6-deposited methylarginine signaling.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
References
De Kouchkovsky I, Abdul-Hay M. ‘Acute myeloid leukemia: a comprehensive review and 2016 update’. Blood Cancer J. 2016;6:e441.
Larrue C, Guiraud N, Mouchel P-L, Dubois M, Farge T, Gotanègre M, et al. Adrenomedullin-CALCRL axis controls relapse-initiating drug tolerant acute myeloid leukemia cells. Nat Commun. 2021;12:422.
van Rhenen A, Feller N, Kelder A, Westra AH, Rombouts E, Zweegman S, et al. High stem cell frequency in acute myeloid leukemia at diagnosis predicts high minimal residual disease and poor survival. Clin Cancer Res. 2005;11:6520–7.
Li X, Yang L, Chen LL. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol Cell. 2018;71:428–42.
Zhou WY, Cai ZR, Liu J, Wang DS, Ju HQ, Xu RH. Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer. 2020;19:172.
Sun YM, Wang WT, Zeng ZC, Chen TQ, Han C, Pan Q, et al. circMYBL2, a circRNA from MYBL2, regulates FLT3 translation by recruiting PTBP1 to promote FLT3-ITD AML progression. Blood. 2019;134:1533–46.
Liu X, Liu X, Cai M, Luo A, He Y, Liu S, et al. CircRNF220, not its linear cognate gene RNF220, regulates cell growth and is associated with relapse in pediatric acute myeloid leukemia. Mol Cancer. 2021;20:139.
Li W, Zhong C, Jiao J, Li P, Cui B, Ji C, et al. Characterization of hsa_circ_0004277 as a New Biomarker for Acute Myeloid Leukemia via Circular RNA Profile and Bioinformatics Analysis. Int J Mol Sci. 2017;18:597.
Han F, Zhong C, Li W, Wang R, Zhang C, Yang X, et al. hsa_circ_0001947 suppresses acute myeloid leukemia progression via targeting hsa-miR-329-5p/CREBRF axis. Epigenomics. 2020;12:935–53.
Yang X, Han F, Hu X, Li G, Wu H, Can C, et al. EIF4A3-induced Circ_0001187 facilitates AML suppression through promoting ubiquitin-proteasomal degradation of METTL3 and decreasing m6A modification level mediated by miR-499a-5p/RNF113A pathway. Biomarker Res. 2023;11:59.
Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV, et al. MLL-Rearranged Leukemia Is Dependent on Aberrant H3K79 Methylation by DOT1L. Cancer Cell. 2011;20:66–78.
Jung N, Dai B, Gentles AJ, Majeti R, Feinberg AP. An LSC epigenetic signature is largely mutation independent and implicates the HOXA cluster in AML pathogenesis. Nat Commun. 2015;6:8489.
He X, Zhu Y, Lin Y-C, Li M, Du J, Dong H, et al. PRMT1-mediated FLT3 arginine methylation promotes maintenance of FLT3-ITD+ acute myeloid leukemia. Blood. 2019;134:548–60.
Tarighat SS, Santhanam R, Frankhouser D, Radomska HS, Lai H, Anghelina M, et al. The dual epigenetic role of PRMT5 in acute myeloid leukemia: gene activation and repression via histone arginine methylation. Leukemia. 2015;30:789–99.
Cheng Y, Gao Z, Zhang T, Wang Y, Xie X, Han G, et al. Decoding m(6)A RNA methylome identifies PRMT6-regulated lipid transport promoting AML stem cell maintenance. Cell Stem Cell. 2023;30:69–85.e7.
Siriboonpiputtana T, Zeisig BB, Zarowiecki M, Fung TK, Mallardo M, Tsai CT, et al. Transcriptional memory of cells of origin overrides β-catenin requirement of MLL cancer stem cells. EMBO J. 2017;36:3139–55.
Chen Z, Gan J, Wei Z, Zhang M, Du Y, Xu C, et al. The Emerging Role of PRMT6 in Cancer. Front Oncol. 2022;12:841381.
Huang T, Yang Y, Song X, Wan X, Wu B, Sastry N, et al. PRMT6 methylation of RCC1 regulates mitosis, tumorigenicity, and radiation response of glioblastoma stem cells. Mol Cell. 2021;81:1276–91.e9.
Wang J, Xiao Z, Li P, Wu C, Li Y, Wang Q, et al. PRMT6-CDC20 facilitates glioblastoma progression via the degradation of CDKN1B. Oncogene. 2023;42:1088–100.
Che N, Ng KY, Wong TL, Tong M, Kau PW, Chan LH, et al. PRMT6 deficiency induces autophagy in hostile microenvironments of hepatocellular carcinoma tumors by regulating BAG5-associated HSC70 stability. Cancer Lett. 2021;501:247–62.
Weng H, Huang H, Wu H, Qin X, Zhao BS, Dong L, et al. METTL14 Inhibits Hematopoietic Stem/Progenitor Differentiation and Promotes Leukemogenesis via mRNA m(6)A Modification. Cell Stem Cell. 2018;22:191–205.e9.
She M, Niu X, Chen X, Li J, Zhou M, He Y, et al. Resistance of leukemic stem-like cells in AML cell line KG1a to natural killer cell-mediated cytotoxicity. Cancer Lett. 2012;318:173–9.
Jang JE, Eom JI, Jeung HK, Chung H, Kim YR, Kim JS, et al. PERK/NRF2 and autophagy form a resistance mechanism against G9a inhibition in leukemia stem cells. J Exp Clin Cancer Res. 2020;39:66.
Gentles AJ, Plevritis SK, Majeti R, Alizadeh AA. Association of a leukemic stem cell gene expression signature with clinical outcomes in acute myeloid leukemia. JAMA. 2010;304:2706–15.
Pellicano F, Park L, Hopcroft LEM, Shah MM, Jackson L, Holyoake TL, et al. hsa-mir183/EGR1-mediated regulation of E2F1 is required for CML stem/progenitor cell survival. Blood. 2018;131:1532–44.
Farge T, Saland E, de Toni F, Aroua N, Hosseini M, Perry R, et al. Chemotherapy-Resistant Human Acute Myeloid Leukemia Cells Are Not Enriched for Leukemic Stem Cells but Require Oxidative Metabolism. Cancer Discov. 2017;7:716–35.
Lagadinou ED, Sach A, Callahan K, Rossi RM, Neering SJ, Minhajuddin M, et al. BCL-2 Inhibition Targets Oxidative Phosphorylation and Selectively Eradicates Quiescent Human Leukemia Stem Cells. Cell Stem Cell. 2013;12:329–41.
Liu J, Wei Y, Jia W, Can C, Wang R, Yang X, et al. Chenodeoxycholic acid suppresses AML progression through promoting lipid peroxidation via ROS/p38 MAPK/DGAT1 pathway and inhibiting M2 macrophage polarization. Redox Biol. 2022;56:102452.
Sébert M, Passet M, Raimbault A, Rahmé R, Raffoux E, Sicre de Fontbrune F, et al. Germline DDX41 mutations define a significant entity within adult MDS/AML patients. Blood. 2019;134:1441–4.
Huang Y, Yu S-H, Zhen W-X, Cheng T, Wang D, Lin J-B, et al. Tanshinone I, a new EZH2 inhibitor restricts normal and malignant hematopoiesis through upregulation of MMP9 and ABCG2. Theranostics. 2021;11:6891–904.
Chen S-J, Bao L, Keefer K, Shanmughapriya S, Chen L, Lee J, et al. Transient receptor potential ion channel TRPM2 promotes AML proliferation and survival through modulation of mitochondrial function, ROS, and autophagy. Cell Death Dis. 2020;11:247.
Reikvam H, Hatfield KJ, Øyan AM, Kalland KH, Kittang AO, Bruserud Ø. Primary human acute myelogenous leukemia cells release matrix metalloproteases and their inhibitors: release profile and pharmacological modulation. Eur J Haematol. 2010;84:239–51.
Kim S, Kim NH, Park JE, Hwang JW, Myung N, Hwang K-T, et al. PRMT6-mediated H3R2me2a guides Aurora B to chromosome arms for proper chromosome segregation. Nat Commun. 2020;11:612.
Cai W, Liu L, Shi X, Liu Y, Wang J, Fang X, et al. Alox15/15-HpETE Aggravates Myocardial Ischemia-Reperfusion Injury by Promoting Cardiomyocyte Ferroptosis. Circulation. 2023;147:1444–60.
Ma X-H, Liu J-H-Z, Liu C-Y, Sun W-Y, Duan W-J, Wang G, et al. ALOX15-launched PUFA-phospholipids peroxidation increases the susceptibility of ferroptosis in ischemia-induced myocardial damage. Signal Transduct Targeted Therapy. 2022;7:288.
Han L, Dong L, Leung K, Zhao Z, Li Y, Gao L, et al. METTL16 drives leukemogenesis and leukemia stem cell self-renewal by reprogramming BCAA metabolism. Cell Stem Cell. 2023;30:52–68.e13.
Li H, Bi K, Feng S, Wang Y, Zhu C. CircRNA circ_POLA2 is Upregulated in Acute Myeloid Leukemia (AML) and Promotes Cell Proliferation by Suppressing the Production of Mature miR-34a. Cancer Manag Res. 2021;13:3629–37.
Hu X, Yang X-Y, Ma D-X. Emerging functions and roles of circRNAs in cancer. Epigenomics. 2022;14:113–6.
de Beauchamp L, Himonas E, Helgason GV. Mitochondrial metabolism as a potential therapeutic target in myeloid leukaemia. Leukemia. 2021;36:1–12.
Amaya ML, Inguva A, Pei S, Jones C, Krug A, Ye H, et al. The STAT3-MYC axis promotes survival of leukemia stem cells by regulating SLC1A5 and oxidative phosphorylation. Blood. 2022;139:584–96.
Herglotz J, Kuvardina ON, Kolodziej S, Kumar A, Hussong H, Grez M, et al. Histone arginine methylation keeps RUNX1 target genes in an intermediate state. Oncogene. 2012;32:2565–75.
Lee J, You JH, Roh J-L. Poly(rC)-binding protein 1 represses ferritinophagy-mediated ferroptosis in head and neck cancer. Redox Biol. 2022;51:102276.
Zhang H, Deng T, Liu R, Ning T, Yang H, Liu D, et al. CAF secreted miR-522 suppresses ferroptosis and promotes acquired chemo-resistance in gastric cancer. Mol Cancer. 2020;19:43.
Acknowledgements
The authors thank all participating patients, without whom continuous improvement in clinical care and growing scientific insights would not be possible. The authors acknowledge all contributing physicians, study nurses, and laboratories for their support in the trials.
Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 32241005, 82370173) and Natural Science Foundation of Shandong Province (Youth Program, ZR2023QH249).
Author information
Authors and Affiliations
Contributions
Professor DXM designed and funded the study. XYY performed this research. JTL, WCL, HYW and CC assisted with this research. YHW, XHJ and XDG analyzed the data. XH, DMW revised the manuscript accordingly. XYY wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The trials were approved by the respective ethics committees and were conducted in accordance with the Declaration of Helsinki. All patients provided written informed consent, including the analysis of data. The studies were continuously monitored using clinical and medical monitoring.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yang, X., Liu, J., Liu, W. et al. circFAM193B interaction with PRMT6 regulates AML leukemia stem cells chemoresistance through altering the oxidative metabolism and lipid peroxidation. Leukemia 38, 1057–1071 (2024). https://doi.org/10.1038/s41375-024-02189-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-024-02189-8