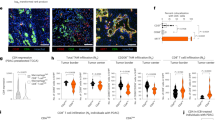
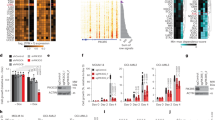
Abstract
Targeting nucleotide biosynthesis is a proven strategy for the treatment of cancer but is limited by toxicity, reflecting the fundamental nucleotide requirement of dividing cells. The rate limiting step in de novo pyrimidine synthesis is of interest, being catalyzed by two homologous enzymes, CTP synthase 1 (CTPS1) and CTPS2, that could be differentially targeted. Herein, analyses of publicly available datasets identified an essential role for CTPS1 in multiple myeloma (MM), linking high expression of CTPS1 (but not CTPS2) with advanced disease and poor outcomes. In cellular experiments, CTPS1 knockout induced apoptosis of MM cell lines. Exposure of MM cells to STP-B, a novel and highly selective pharmacological inhibitor of CTPS1, inhibited proliferation, induced S phase arrest and led to cell death by apoptosis. Mechanistically, CTPS1 inhibition by STP-B activated DNA damage response (DDR) pathways and induced double-strand DNA breaks which accumulated in early S phase. Combination of STP-B with pharmacological inhibitors of key components of the DDR pathway (ATR, CHEK1 or WEE1) resulted in synergistic growth inhibition and early apoptosis. Taken together, these findings identify CTPS1 as a promising new target in MM, either alone or in combination with DDR pathway inhibition.

This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Szalat R, Munshi NC. Novel Agents in Multiple Myeloma. Cancer J. 2019;25:45–53.
Zanwar S, Kumar S. Disease heterogeneity, prognostication and the role of targeted therapy in multiple myeloma. Leuk Lymphoma. 2021;62:3087–97.
Pawlyn C, Davies FE. Toward personalized treatment in multiple myeloma based on molecular characteristics. Blood. 2019;133:660–75.
Weinhold N, Heuck CJ, Rosenthal A, Thanendrarajan S, Stein CK, Van Rhee F, et al. Clinical value of molecular subtyping multiple myeloma using gene expression profiling. Leukemia. 2016;30:423–30.
Kumar SK, Lee JH, Lahuerta JJ, Morgan G, Richardson PG, Crowley J, et al. Risk of progression and survival in multiple myeloma relapsing after therapy with IMiDs and bortezomib: a multicenter international myeloma working group study. Leukemia. 2012;26:149–57.
Kumar SK, Dimopoulos MA, Kastritis E, Terpos E, Nahi H, Goldschmidt H, et al. Natural history of relapsed myeloma, refractory to immunomodulatory drugs and proteasome inhibitors: a multicenter IMWG study. Leukemia. 2017;31:2443.
Moore M, Maroun J, Robert F, Natale R, Neidhart J, Dallaire B, et al. Multicenter phase II study of brequinar sodium in patients with advanced gastrointestinal cancer. Invest New Drugs. 1993;11:61–65.
Urba S, Doroshow J, Cripps C, Robert F, Velez-Garcia E, Dallaire B, et al. Multicenter phase II trial of brequinar sodium in patients with advanced squamous-cell carcinoma of the head and neck. Cancer Chemother Pharmacol. 1992;31:167–9.
Dodion PF, Wagener T, Stoter G, Drozd A, Lev LM, Skovsgaard T, et al. Phase II trial with Brequinar (DUP-785, NSC 368390) in patients with metastatic colorectal cancer: a study of the Early Clinical Trials Group of the EORTC. Ann Oncol Off J Eur Soc Med Oncol. 1990;1:79–80.
Zhou Y, Tao L, Zhou X, Zuo Z, Gong J, Liu X, et al. DHODH and cancer: promising prospects to be explored. Cancer Metab. 2021;9:22.
Evans DR, Guy HI. Mammalian pyrimidine biosynthesis: fresh insights into an ancient pathway. J Biol Chem. 2004;279:33035–8.
Lieberman I. Enzymatic amination of uridine triphosphate to cytidine triphosphate. J Biol Chem. 1956;222:765–75.
Martin E, Palmic N, Sanquer S, Lenoir C, Hauck F, Mongellaz C, et al. CTP synthase 1 deficiency in humans reveals its central role in lymphocyte proliferation. Nature. 2014;510:288.
Martin E, Minet N, Boschat A-C, Sanquer S, Sobrino S, Lenoir C, et al. Impaired lymphocyte function and differentiation in CTPS1-deficient patients result from a hypomorphic homozygous mutation. JCI insight. 2020;5:e133880.
Lynch EM, DiMattia MA, Albanese S, van Zundert GCP, Hansen JM, Quispe JD, et al. Structural basis for isoform-specific inhibition of human CTPS1. Proc Natl Acad Sci. 2021;118:e2107968118.
Asnagli H, Minet N, Pfeiffer C, Hoeben E, Lane R, Laughton D, et al. CTP Synthase 1 Is a Novel Therapeutic Target in Lymphoma. HemaSphere. 2023;7:e864.
Altman DG, Lausen B, Sauerbrei W, Schumacher M. Dangers of using ‘optimal’ cutpoints in the evaluation of prognostic factors. J Natl Cancer Inst. 1994;86:829–35.
Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, et al. Defining a Cancer Dependency Map. Cell. 2017;170:564–576.e16.
Dempster JM, Boyle I, Vazquez F, Root DE, Boehm JS, Hahn WC, et al. Chronos: a cell population dynamics model of CRISPR experiments that improves inference of gene fitness effects. Genome Biol. 2021;22:343.
Agnelli L, Mosca L, Fabris S, Lionetti M, Andronache A, Kwee I, et al. A SNP microarray and FISH-based procedure to detect allelic imbalances in multiple myeloma: an integrated genomics approach reveals a wide gene dosage effect. Genes Chromosomes Cancer. 2009;48:603–14.
Chng WJ, Kumar S, Vanwier S, Ahmann G, Price-Troska T, Henderson K, et al. Molecular dissection of hyperdiploid multiple myeloma by gene expression profiling. Cancer Res. 2007;67:2982–9.
Broyl A, Hose D, Lokhorst H, de Knegt Y, Peeters J, Jauch A, et al. Gene expression profiling for molecular classification of multiple myeloma in newly diagnosed patients. Blood. 2010;116:2543–53.
Zhan F, Huang Y, Colla S, Stewart JP, Hanamura I, Gupta S, et al. The molecular classification of multiple myeloma. Blood. 2006;108:2020–8.
Hanamura I, Huang Y, Zhan F, Barlogie B, Shaughnessy J. Prognostic value of Cyclin D2 mRNA expression in newly diagnosed multiple myeloma treated with high-dose chemotherapy and tandem autologous stem cell transplantations. Leukemia. 2006;20:1288–90.
Mulligan G, Mitsiades C, Bryant B, Zhan F, Chng WJ, Roels S, et al. Gene expression profiling and correlation with outcome in clinical trials of the proteasome inhibitor bortezomib. Blood. 2007;109:3177–88.
Traut TW. Physiological concentrations of purines and pyrimidines. Mol Cell Biochem. 1994;140:1–22.
Kawano Y, Moschetta M, Manier S, Glavey S, Görgün GT, Roccaro AM, et al. Targeting the bone marrow microenvironment in multiple myeloma. Immunol Rev. 2015;263:160–72.
Pelletier J, Thomas G, Volarević S. Ribosome biogenesis in cancer: new players and therapeutic avenues. Nat Rev Cancer. 2018;18:51–63.
Matthews HK, Bertoli C, de Bruin RAM. Cell cycle control in cancer. Nat Rev Mol Cell Biol. 2022;23:74–88.
Huang H, Wang Y, Wang W, Wei X, Gale RP, Li J, et al. A prognostic survival model based on metabolism-related gene expression in plasma cell myeloma. Leukemia. 2021;35:3212–22.
Ajazi A, Choudhary R, Tronci L, Bachi A, Bruhn C. CTP sensing and Mec1ATR-Rad53CHK1/CHK2 mediate a two-layered response to inhibition of glutamine metabolism. PLoS Genet. 2022;18:e1010101.
Sun Z, Zhang Z, Wang Q-Q, Liu J-L. Combined Inactivation of CTPS1 and ATR Is Synthetically Lethal to MYC-Overexpressing Cancer Cells. Cancer Res. 2022;82:1013–24.
Walters DK, Wu X, Tschumper RC, Arendt BK, Huddleston PM, Henderson KJ, et al. Evidence for ongoing DNA damage in multiple myeloma cells as revealed by constitutive phosphorylation of H2AX. Leukemia. 2011;25:1344–53.
Cottini F, Hideshima T, Xu C, Sattler M, Dori M, Agnelli L, et al. Rescue of Hippo coactivator YAP1 triggers DNA damage-induced apoptosis in hematological cancers. Nat Med. 2014;20:599–606.
Cottini F, Hideshima T, Suzuki R, Tai Y-T, Bianchini G, Richardson PG, et al. Synthetic Lethal Approaches Exploiting DNA Damage in Aggressive Myeloma. Cancer Discov. 2015;5:972–87.
Herrero AB, Gutiérrez NC. Targeting Ongoing DNA Damage in Multiple Myeloma: Effects of DNA Damage Response Inhibitors on Plasma Cell Survival. Front Oncol. 2017;7:98.
Botrugno OA, Bianchessi S, Zambroni D, Frenquelli M, Belloni D, Bongiovanni L, et al. ATR addiction in multiple myeloma: synthetic lethal approaches exploiting established therapies. Haematologica. 2020;105:2440–7.
Shukla SK, Purohit V, Mehla K, Gunda V, Chaika NV, Vernucci E, et al. MUC1 and HIF-1alpha Signaling Crosstalk Induces Anabolic Glucose Metabolism to Impart Gemcitabine Resistance to Pancreatic Cancer. Cancer Cell. 2017;32:71–87.
Conte F, Sibilio P, Grimaldi AM, Salvatore M, Paci P, Incoronato M. In silico recognition of a prognostic signature in basal-like breast cancer patients. PLoS One. 2022;17:e0264024.
Lin Y, Zhang J, Li Y, Guo W, Chen L, Chen M, et al. CTPS1 promotes malignant progression of triple-negative breast cancer with transcriptional activation by YBX1. J Transl Med. 2022;20:17.
Minet N, Boschat A-C, Lane R, Laughton D, Beer P, Asnagli H, et al. Differential roles of CTP synthetases CTPS1 and CTPS2 in cell proliferation. Life Sci Alliance. 2023;6:e202302066.
Acknowledgements
The authors would like to thank Waltraud Scherbler and Pia Fritz (employees at Klinik Ottakring, Vienna, Austria), for their excellent technical support. This study was supported by the Austrian Forum against Cancer, the Österreichische Forschungsförderungsgesellschaft mbH (FFG, “Forschungspartnerschaften”), project number 878861 and a research grant from the Ingrid Shaker Nessmann (ISNK) Cancer Research Association given to AB; AS is a recipient of the DOC fellowship of the Austrian Academy of Science at the Wilhelminen Cancer Research Institute. In vivo studies conducted at Crown Bioscience were financed by Step Pharma.
Author information
Authors and Affiliations
Contributions
CP, AB, PAB and HL designed the study; CP performed the experiments, analyzed and interpreted data; AMG performed bioinformatics analysis of publicly available gene expression datasets; AB, AS, AMG, AEP, HA, JH, and PAB analyzed and interpreted data; NZ and MS interpreted data; CP and PAB. wrote the manuscript; HL provided funding, designed the study and supervised the research; all authors revised and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
Philip A. Beer, Hélène Asnagli and Andrew E. Parker are employees of Step Pharma SAS. All other authors declare no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pfeiffer, C., Grandits, A.M., Asnagli, H. et al. CTPS1 is a novel therapeutic target in multiple myeloma which synergizes with inhibition of CHEK1, ATR or WEE1. Leukemia 38, 181–192 (2024). https://doi.org/10.1038/s41375-023-02071-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-023-02071-z
This article is cited by
-
Inactivation of cytidine triphosphate synthase 1 prevents fatal auto-immunity in mice
Nature Communications (2024)