Abstract
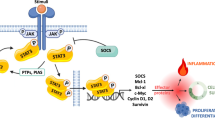
LGL disorders are rare hematological neoplasias with remarkable phenotypic, genotypic and clinical heterogeneity. Despite these constraints, many achievements have been recently accomplished in understanding the aberrant pathways involved in the LGL leukemogenesis. In particular, compelling evidence implicates STAT signaling as a crucial player of the abnormal cell survival. As interest increases in mapping hematological malignancies by molecular genetics, the relevance of STAT gene mutations in LGL disorders has emerged thanks to their association with discrete clinical features. STAT3 and STAT5b mutations are recognized as the most common gain-of-function genetic lesions up to now identified in T-LGL leukemia (T-LGLL) and are actually regarded as the hallmark of this disorder, also contributing to further refine its subclassification. However, from a clinical perspective, the relationships between T-LGLL and other borderline and overlapping conditions, including reactive cell expansions, clonal hematopoiesis of indeterminate potential (CHIP) and unrelated clonopathies are not fully established, sometimes making the diagnosis of T cell malignancy challenging. In this review specifically focused on the topic of clonality of T-LGL disorders we will discuss the rationale of the appropriate steps to aid in distinguishing LGLL from its mimics, also attempting to provide new clues to stimulate further investigations designed to move this field forward.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Loughran TP Jr, Kadin ME, Starkebaum G, Abkowitz JL, Clark EA, Disteche C, et al. Leukemia of large granular lymphocytes: association with clonal chromosomal abnormalities and autoimmune neutropenia, thrombocytopenia, and hemolytic anemia. Ann Intern Med. 1985;102:169–75.
Semenzato G, Pandolfi F, Chisesi T, De Rossi G, Pizzolo G, Zambello R, et al. The lymphoproliferative disease of granular lymphocytes. A heterogeneous disease ranging from indolent to aggressive conditions. Cancer. 1987;60:2971–8.
Oshimi K, Yamada O, Kaneko T, Nishinarita S, Izuka Y, Urabe A, et al. Laboratory findings and clinical courses of 33 patients with granular lymphocyte-proliferative disorders. Leuk (Baltim). 1993;7:782–8.
Zambello R, Semenzato G. Large granular lymphocyte disorders: new etiopathogenetic clues as a rationale for innovative therapeutic approaches. Haematologica. 2009;94:1341–5.
Lamy T, Moignet A, Loughran TP Jr. LGL leukemia: from pathogenesis to treatment. Blood. 2017;129:1082–94.
Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon, France: IARC Press; 2017.
Alaggio R, Amador C, Anagnostopoulos I, Attygalle AD, Araujo IB de O, Berti E, et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Lymphoid Neoplasms. Leukemia. 2022;36:1720–48.
Semenzato G, Pizzolo G, Ranucci A, Agostini C, Chilosi M, Quinti I, et al. Abnormal expansions of polyclonal large to small size granular lymphocytes: reactive or neoplastic process? Blood. 1984;63:1271–7.
Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334:395–402.
Lima M, Almeida J, Santos AH, dos Anjos Teixeira M, Alguero MC, Queiros ML, et al. Immunophenotypic analysis of the TCR-Vbeta repertoire in 98 persistent expansions of CD3(+)/TCR-alphabeta(+) large granular lymphocytes: utility in assessing clonality and insights into the pathogenesis of the disease. Am J Pathol. 2001;159:1861–8.
Langerak AW, van Den Beemd R, Wolvers-Tettero ILM, Boor PP, van Lochem EG, Hooljkaas H, et al. Molecular and flow cytometric analysis of the Vbeta repertoire for clonality assessment in mature TCRalphabeta T-cell proliferations. Blood. 2001;98:165–73.
Shi M, Jevremivic D, Otteson GE, Timm MM, OLteanu H, Horna P. Single antibody detection of T-cell receptor ab clonality by flow cytometry rapidly identifies mature T-cell neoplasms and monotypic small CD8-positive subsets of uncertain significance. Cytom Part B. 2020;98B:99–107.
Munoz-Garcia N, Moran-Plata FJ, Villamor N, Lima M, Barrena S, Mateos S, et al. High-sensitive TRBC1-based flow cytometric assessment of T-cell clonality in Tαβ-large granular lymphocytic leukemia. Cancers. 2022;14:408–17.
Clemente MJ, Wlodarski MW, Makishima H, Viny AD, Bretschneider I, Shaik M, et al. Clonal drift demonstrates unexpected dynamics of the T-cell repertoire in T-large granular lymphocyte leukemia. Blood. 2011;118:4384–93.
Gattazzo C, Teramo A, Passeri F, De March E, Carraro S, Trimarco V, et al. Detection of monoclonal T cell populations in patients with KIR-restricted chronic lymphoproliferative disorder of NK cells. Haematologica. 2014;99:1826–33.
Semenzato G, Zambello R, Starkebaum G, Oshimi K, Loughran TP Jr. The lymphoproliferative disease of granular lymphocytes: updated criteria for the diagnosis. Blood. 1997;89:256–60.
Barcena P, Jara-Acevedo M, Tabernero MD, Lopez A, Sanchez ML, Garcia-Montero AC, et al. Phenoptypic profile of expanded NK cells in chronic lymphoproliferative disorders: a surrogate marker for NK-cell clonality. Oncotarget. 2015;6:42938–52.
Zambello R, Falco M, Della Chiesa M, Trentin L, Carollo D, Castriconi R, et al. Expression and function of KIR and natural cytotoxicity receptors in NK-type lymphoproliferative diseases of granular lymphocytes. Blood. 2003;102:1797–805.
Morice WG, Kurtin PJ, Tefferi A, Hanson CA. Distinct bone marrow findings in T-cell granular lymphocytic leukemia revealed by paraffin section immunoperoxidase stains for CD8, TIA-1, and granzyme B. Blood. 2002;99:268–74.
Osuji N, Beiske K, Randen U, Matutes E, Tjonnfjord G, Catovsky D, et al. Characteristic appearances of the bone marrow in T-cell large granular lymphocyte leukaemia. Histopathology. 2007;50:547–74.
Mustjoki S, Young NS. Somatic mutations in “benign” disease. N. Engl J Med. 2021;384:2039–52.
Busque L, Patel JP, Figueroa ME, Vasanthakumar A, Provost S, Hamilou Z, et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat Genet. 2012;44:1179–81.
Genovese G, Kahler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl J Med. 2014;371:2477–87.
Jaiswal S, Ebert BL. Clonal hematopoiesis in human aging and disease. Science. 2019;366:6465.
Steensma DP, Bejar R, Jaiswal S, Lindsley RC, Sekeres MA, Hasserjian RP, et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126:9–16.
Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, et al. Age-related clonal hematopoiesis associated with adverse outcome. N. Engl J Med. 2014;371:2488–98.
Rossi M, Meggendorfer M, Zampini M, Tettamanti M, Riva E, Travaglino E, et al. Clinical relevance of clonal hematopoiesis in persons aged >80 years. Blood. 2021;138:2093–105.
Razavi P, Li BT, Brown DN, Jung B, Hubbel E, Shen R, et al. High intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat Med. 2019;25:1928–37.
Yizhak K, Aguet F, Kim J, Hess JM, Kubler K, Grimsby J, et al. RNAsequence analysis reveals macroscopic somatic clonal expansion across normal tissues. Science. 2019;364(6444):eaaw0726.
Young AL, Challen GA, Birmann BM, Druley TE. Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults. Nat Commun. 2016;7:12484.
Fuster JJ, MacLauchlan S, Zuriaga MA, Polackal MN, Ostriker AC, Chakraborty R, et al. Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science. 2017;355:842–7.
Abegunde SO, Buckstein R, Wells RA, Rauh MJ. An inflammatory environment containing TNFα favors Tet2-mutant clonal hematopoiesis. Exp Hematol. 2018;59:60–65.
Florez MA, Tran TB, Wathan TK, DeGregory J, Pietras EM, King KY. Clonal hematopoiesis: Mutation-specific adaptation to environmental change. Cell Stem Cells. 2022;29:882–904.
Watson CJ, Papula AL, Poon GYP, Wong WH, Young AL, Druley TE, et al. The evolutionary dynamics and fitness landscape of clonal hematopoiesis. Science. 2020;367:1449–54.
Gorodetskiy VR, Sidorova YV, Kupryshina NA, Vasilyev VI, Probatova NA, Ryzhikova NV, et al. Analysis of a single-institution cohort of patients with Felty’s syndrome and T-cell large granular lymphocyte leukemia in the setting of rheumatoid arthritis. Reum Intern. 2021;41:147–56.
Dameshek W, Schwartz RS. Leukemia and auto-immunization-some possible relationships. Blood. 1959;14:1151–8.
Komrokji RS, al Ali N, Sallman D, Padron E, Lancet J, Sokol L, et al. Characterization of myelodysplastic syndromes (MDS) with T-cell large granular lymphocyte proliferations (LGL). Leukemia. 2020;34:3097–9.
Durrani J, Awada H, Kishtagari A, Visconti V, Kerr K, Adema V, et al. Large granular lymphocytic leukemia coexists with myeloid clones and myelodysplastic syndrome. Leukemia. 2020;34:957–62.
Calabretto G, Attardi E, Teramo A, Trimarco V, Carraro S, Mossuto S, et al. An Italian multi-center experience of hypocellular Myelodysplastic Syndromes (h-MDS): from clinical description to immunological characterization. Leukemia. 2022;36:1947–50.
Jerez A, Clemente MJ, Makishima H, Rajala H, Gòmez-Seguì I, Olson T, et al. STAT3 mutations indicate the presence of subclinical T-cell clones in a subset of aplastic anemia and myelodysplastic syndrome patients. Blood. 2013;122:2453–9.
Lundgren S, Keranen MAI, Kankainen M, Huuhtanen J, Walldin G, Kerr CM, et al. Somatic mutations in lymphocytes in patients with immune-mediated aplastic anemia. Leukemia. 2021;35:1365–79.
Kawakami F, Kawakami T, Ymane T, Maruyama M, Kobayashi J, Nishima S, et al. T cell clonal expansion and STAT3 mutations: a characteristic feature of acquired chronic T cell-mediated pure red cell aplasia. Int J Hematol. 2022;115:816–25.
Zhang R, Shah MV, Loughran TP. The root of many evils: indolent large granular lymphocyte leukaemia and associated disorders. Hematol Oncol. 2010;28:105–17.
Kreutzman A, Juvonen V, Kairisto V, Ekblom M, Stenke L, Seggewiss R, et al. Mono/oligoclonal T and NK cells are common in chronic myeloid leukemia patients at diagnosis and expand during dasatinib therapy. Blood. 2010;116:772–82.
Risitano AM, Maciejewski JP, Muranski P, Wlodarski M, O’Keefe C, Sloand EM, et al. Large granular lymphocyte (LGL)-like clonal expansions in paroxysmal nocturnal hemoglobinuria (PNH) patients. Leukemia. 2005;19:217–22.
Munoz-Ballester J, Chen-Liang TH, Hurtado AM, Heras I, de Arriba F, Garcìa-Maio MD, et al. Persistent cytotoxic lymphocyte expansions after allogenic haematopoietic stem cell transplantation: kinetics, clinical impact and absence of STAT3 mutations. Br J Haemat. 2016;172:937–46.
Miller PG, Qiao D, Rojas-Quintero J, Honigberg MC, Miller PG, Qiao D, et al. Association of clonal hematopoiesis with chronic obstructive pulmonary disease. Blood. 2022;139:357–69.
Dhodapkar MV, Li C-Y, Lust JA, Phylik RL. Clinical Spectrum of Clonal Proliferations of T-Large Granular Lymphocytes: A T-cell Clonopathy of Undetermined Significance? Blood. 1984;84:1620–7.
Shi M, Olteanu H, Jevremovic D, He R, Viswanatha D, Corley H, et al. T-cell clones of uncertain significance are highly prevalent and show close resemblance to T-cell large granular lymphocytic leukemia. Implic Lab diagnostics Mod Pathol. 2020;33:2046–57.
Sabnani I, Zucker MJ, Tsang P, Palekar S. Clonal T-large granular lymphocyte proliferation in solid organ transplant recipients. Transpl Proc. 2006;38:3437–40.
Taylor HG, Terebelo HR, Gamez A. Lymphocytosis in a patient with malignant fibrous histiocytoma. Cancer. 1982;50:1563–7.
Zambello R, Loughran TP Jr, Trentin L, Rassu M, Facco M, Bortolin M, et al. Spontaneous resolution of p58/EB6 antigen restricted NK-type lymphoproliferative disease of granular lymphocytes: role of Epstein Barr virus infection. Br J Haematol. 1997;99:215–21.
Venturi V, Price DA, Douek DC, Davemport MP. The molecular basis for public T-cell responses? Nat Rev Immunol. 2008;8:231–8.
Emerson RO, DeWitt WS, Vignali M, Gravley J, Hu JK, Osborne EJ, et al. Immunosequencing identifies signatures of cytomegalovirus exposure history and HLA-mediated effects on the T cell repertoire. Nat Genet. 2017;49:659–65.
Miles JJ, Douek DC, Price DA. Bias in the αβ T-cell repertoire: implications for disease pathogenesis and vaccination. Immuno Cell Biol. 2011;89:375–87.
Bhattacharya D, Teramo A, Gasparini VR, HuuhtanenJ, Kim D, Theodoropoulos J, et al. Identification of novel STAT5B mutations and characterization of TCRβ signatures in CD4+ T large granular lymphocyte leukemia. Blood Cancer J. 2022;12:31–42.
Clemente MJ, Przychodzen B, Jerez A, Dienes BE, Afable MG, Husseinzadeh H, et al. Deep sequencing of the T-cell receptor repertoire in CD8+ T-large granular lymphocyte leukemia identifies signature landscapes. Blood. 2013;122:4077–85.
Garrido P, Ruiz-Cabello F, Barcena P, Sandberg Y, Cantòn J, Lima M, et al. Monoclonal TCR-Vbeta13.1+/CD4+/NKa+/CD8−/+dim T-LGL lymphocytosis: evidence for an antigen-driven chronic T-cell stimulation origin. Blood. 2007;109:4890–8.
Klein K, Stoiber D, Sexl V, Witalisz-Siepracka A. Untwining anti-tumor and immunosuppressive effects of JAK inhibitors. A strategy of hematological malignancies? Cancers. 2021;13:2611–35.
Kim D, Myllymaki M, Kankainen M, Jarvinen T, Park G, Bruhn R, et al. Somatic STAT mutations in CD8+ T cells of healthy blood donors carrying human T-cell leukemia virus type 2. Haematologica. 2022;107:550–4.
Koskela HL, Eldfors S, Ellonen P, Adrichem AJ, Kuusanmaki H, Andersson EI, et al. Somatic STAT3 mutations in large granular lymphocytic leukemia. N. Engl J Med. 2012;366:1905–13.
Andersson EI, Tanahashi T, Sekiguchi N, Gasparini VR, Bortoluzzi S, Kawakami T, et al. High incidence of activating STAT5B mutations in CD4-positive T-cell large granular lymphocyte leukemia. Blood. 2016;128:2465–8.
Teramo A, Barilà G, Calabretto G, Ercolin C, Moignet A, Roussel M, et al. STAT3 mutation impacts biological and clinical features of T-LGL leukemia. Oncotarget. 2017;8:61876–89.
Shi M, He R, Feldman AL, Viswanatha DS, Jeremovic D, Chen D, et al. STAT3 mutations and its clinical and histopathologic correlation in T-cell large granular lymphocytic leukemia. Hum Pathol. 2018;73:74–81.
Barilà G, Teramo A, Calabretto G, Vicenzetto C, Gasparini VR, Pavan L, et al. Stat3 mutations impact on overall survival in large granular lymphocyte leukemia: a single-center experience of 205 patients. Leukemia. 2020;34:1116–24.
Muñoz-Garcìa N, Jara-Acevedo M, Caldas C, Barcena P, Lòpez A, Puig N, et al. STAT3 and STAT5B mutations in T/NK-chronic lymphoproliferative disorders of large granular lymphocytes (LGL): association with disease features. Cancers. 2020;12:3508–30.
Rajala HLM, Eldfors S, Kuusanmaki H, van Adrichem AJ, Olson T, Lagstrom S, et al. Discovery of somatic STAT5b mutations in large granular lymphocyte leukemia. Blood. 2013;121:4541–50.
Calabretto G, Teramo A, Barilà G, Vicenzetto C, Gasparetto VR, Semenzato G, et al. Neutropenia and Large Granular Lymphocyte Leukemia: from pathogenesis to therapeutic options. Cells. 2021;10:2800–15.
Teramo A, Binatti A, Ciabatti E, Schiavoni G, Tarrini G, Barilà G, et al. Defining TCRγδ lymphoproliferative disorders by combined immunophenotypic and molecular evaluation. Nat Comm. 2022;13:3298–308.
Barilà G, Grassi A, Cheon HJ, Teramo A, Calabretto G, Chahal J et al. Tγδ large granular lymphocyte leukemia identifies a subset of patients with more symptomatic disease: analysis of a large international cohort of 127 patients. Blood. 2022; In press.
Epling-Burnette PK, Liu JH, Catlett-Falcone R, Turkson J, Oshiro M, Kothapalli R, et al. Inhibition of STAT3 signaling leads to apoptosis of leukemic large granular lymphocytes and decreased Mcl-1 expression. J Clin Invest. 2001;107:351–62.
Teramo A, Gattazzo C, Passeri F, Lico A, Tasca G, Cabrelle A, et al. Intrinsic and extrinsic mechanisms contribute to maintain the JAK/STAT pathway aberrantly activated in T-type large granular lymphocyte leukemia. Blood. 2013;121:3843–54.
Yu H, Jove R. The STATs of cancer-new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105.
Kim D, Park G, Huuhtanen J, Ghimire B, Rajala H, Moriggi R, et al. STAT3 activation in large granular lymphocyte leukemia is associated with cytokine signaling and DNA hypermethylation. Leukemia. 2021;35:3430–43.
Mariotti B, Calabretto G, Rossato M, Teramo A, Castellucci M, Barilà G, et al. Identification of a miR-146b-FasL axis in the development of neutropenia in T-Large Granular Lymphocyte Leukemia. Haematologica. 2020;105:2040–52.
Dutta A, Yan D, Hutchinson RE, Mohi G. Stat3 mutations are not sufficient to induce large granular lymphocytic leukaemia in mice. Br J Haematol. 2018;180:911–5.
Kerr CM, Clemente MJ, Chomoczynski PW, Przychodzen B, Nagata Y, Adema V, et al. Subclonal STAT mutations solidify clonal dominance. Blood Adv. 2019;3:917–21.
Coppe A, Andersson EI, Binatti A, Gasparini VR, Bortoluzzi S, Clemente M, et al. Genomic landscape characterization of large granular lymphocyte leukemia with a systems genetics approach. Leukemia. 2017;31:1243–6.
Cheon H, Xing JC, Moosic KB, Ung J, Chan VW, Chung DS, et al. Genomic and transcriptomic sequencing reveals the landscape of large granular lymphocyte leukemia. Blood. 2022;139:3058–72.
Teramo A, Barilà G, Calabretto G, Vicenzetto C, Gasparini VR, Semenzato G, et al. Insights into genetic landscape of large granular lymphocyte leukemia. Front Oncol. 2020;10:152–9.
Raess PW, Cascio MJ, Fan G, Press R, Druker BJ, Brewer D, et al. Concurrent STAT3, DNMT3A, and TET2 mutations in T-LGL leukemia with molecularly distinct clonal hematopoiesis of indeterminate potential. Am J Hematol. 2017;92:E6–E8.
Assmann JLJC, Leon LG, Stavast CJ, van den Bogaerdt SE, Schilperoord J, Sandberg Y, et al. miR-181a is a novel payer in the STAT3-mediated survival network of TCRαβ+ CD8+ T large granular lymphocyte leukemia. Leukemia. 2022;36:1480–2.
Loughran TP, Zicki L, Olson TL, Wang V, Zhang D, Rajala HLM, et al. Immunosuppressive Therapy of LGL Leukemia: Prospective multicenter phase II study by the Eastern Cooperative Oncology Group (E5998). Leukemia. 2015;29:886–94.
Semenzato G, Zambello R. Interrogating molecular genetics to refine LGLL classification. Blood. 2022;139:3002–4.
Moskowitz AJ, Ghione P, Jacobsen E, Ruan J, Schatz JH, Noor S, et al. Phase 2 biomarker-driven study of ruxolitinib demonstrates effectiveness of JAK/STAT targeting in T-cell lymphomas. Blood. 2021;138:2827–36.
Zambello R, Berno T, Cannas G, Baesso I, Binotto G, Bonoldi E, et al. Phenotypic and functional analyses of dendritic cells in patients with lymphoproliferative disease of granular lymphocytes (LDGL). Blood. 2005;106:3926–31.
Andersson EI, Pützer S, Yadav B, Dufva O, Khan S, He L, et al. Discovery of novel drug sensitivities in T-PLL by high-throughput ex vivo drug testing and mutation profiling. Leukemia. 2018;32:774–7.
Huuhtanen J, Bhattacharya D, Lonnberg T, Kankainen M, Kerr C, Theodoropoulos J, et al. Single-cell transcriptomics identifies synergistic role of leukemic and non-leukemic immune repertories in CD8+ T cell Large Granular Lymphocytic Leukemia. Nat Commun. 2022;13:1981–96.
Gao S, Wu Z, Arnold B, Diamond C, Batchu S, Giudice V, et al. Single-cell RNA sequencing coupled to TCR profiling of large granular lymphocyte leukemia T cells. Nat Commun. 2022;13:1982–94.
Fasan A, Kern W, Grossmann V, Haferlach C, Haferlach S, Schnittger S. STAT3 mutations are highly specific for large granular lymphocytic leukemia. Leukemia. 2013;27:1598–1600.
Acknowledgements
This work has been supported by Associazione Italiana per la Ricerca sul Cancro (AIRC, IG-20216 to GS).
Author information
Authors and Affiliations
Contributions
GS contributed to the conceptualization of the Review, GS, AT, GC, VRG, RZ to the literature review and data collection. GS, AT, GC, RZ contributed to the figures. GS and RZ contributed to writing the original draft. GS, AT, GC, VRG, RZ contributed to review and edit subsequent drafts.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Semenzato, G., Teramo, A., Calabretto, G. et al. All that glitters is not LGL Leukemia. Leukemia 36, 2551–2557 (2022). https://doi.org/10.1038/s41375-022-01695-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41375-022-01695-x
This article is cited by
-
The constitutive activation of STAT3 gene and its mutations are at the crossroad between LGL leukemia and autoimmune disorders
Blood Cancer Journal (2024)
-
T-large granular lymphocyte frequencies and correlates in disease states detected by multiparameter flow cytometry in pediatric and young adult population
Annals of Hematology (2024)