Abstract
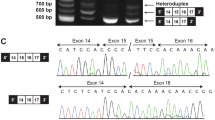
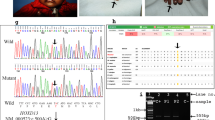
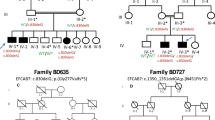
Cenani-Lenz syndrome (CLS) is a rare autosomal-recessive congenital disorder affecting development of distal limbs. It is characterized mainly by syndactyly and/or oligodactyly, renal anomalies, and characteristic facial features. Mutations in the LRP4 gene, located on human chromosome 11p11.2–q13.1, causes the CLS. The gene LRP4 encodes a low-density lipoprotein receptor-related protein-4, which mediates SOST-dependent inhibition of bone formation and Wnt signaling. In the study, presented here, three families of Pakistani origin, segregating CLS in the autosomal recessive manner were clinically and genetically characterized. In two families (A and B), microsatellite-based homozygosity mapping followed by Sanger sequencing identified a novel homozygous missense variant [NM_002334.3: c.295G>C; p.(Asp99His)] in the LRP4 gene. In the third family C, exome sequencing revealed a second novel homozygous missense variant [NM_002334.3: c.1633C>T; p.(Arg545Trp)] in the same gene. To determine the functional relevance of these variants, we tested their ability to inhibit canonical WNT signaling in a luciferase assay. Wild type LRP4 was able to inhibit LRP6-dependent WNT signaling robustly. The two mutants p.(Asp99His) and p.(Arg545Trp) inhibited WNT signaling less effectively, suggesting they reduced LRP4 function.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Tian J, Shao J, Liu C, Hou HY, Chou CW, Shboul M, et al. Deficiency of lrp4 in zebrafish and human LRP4 mutation induce aberrant activation of Jagged–Notch signaling in fin and limb development. Cell Mol Life Sci. 2019;76:163–78.
Li Y, Pawlik B, Elcioglu N, Aglan M, Kayserili H, Yigit G, et al. LRP4 mutations alter Wnt/β-catenin signaling and cause limb and kidney malformations in Cenani-Lenz syndrome. Am J Hum Genet. 2010;86:696–706.
Ohkawara B, Cabrera-Serrano M, Nakata T, Milone M, Asai N, Ito K, et al. LRP4 third β-propeller domain mutations cause novel congenital myasthenia by compromising agrin-mediated MuSK signaling in a position-specific manner. Hum Mol Genet. 2014;23:1856–68.
Xiong L, Jung JU, Wu H, Xia WF, Pan JX, Shen C, et al. Lrp4 in osteoblasts suppresses bone formation and promotes osteoclastogenesis and bone resorption. Nat Acad Sci. 2015;112:3487–92.
Dietrich MF, Van Der Weyden L, Prosser HM, Bradley A, Herz J, Adams DJ. Ectodomains of the LDL receptor-related proteins LRP1b and LRP4 have anchorage independent functions in vivo. PLoS ONE. 2010;5:e9960.
Johnson EB, Hammer RE, Herz J. Abnormal development of the apical ectodermal ridge and polysyndactyly in Megf7-deficient mice. Hum Mol Genet. 2005;14:3523–38.
Johnson EB, Steffen DJ, Lynch KW, Herz J. Defective splicing of Megf7/Lrp4, a regulator of distal limb development, in autosomal recessive mulefoot disease. Genomics 2006;88:600–9.
Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. In: Bioinform Methods Prot. 2000:365–86.
Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009;25:1754–60.
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Gen Res. 2010;20:1297–303.
Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucl Acid Res. 2010;38:164.
Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91.
Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet.2014;46:310–5.
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comp Chem. 2004;25:1605–12.
Nakayama M, Nakajima D, Nagase T, Nomura N, Seki N, Ohara O. Identification of high-molecular-weight proteins with multiple EGF-like motifs by motif-trap screening. Genomics. 1998;51:27–34.
Cenani A, Lenz W. Total syndactyly and total radio-ulnar synostosis in 2 brothers. Zeit Fur Kind. 1967;101:181.
Leupin O, Piters E, Halleux C, Hu S, Kramer I, Morvan F, et al. Bone overgrowth-associated mutations in the LRP4 gene impair sclerostin facilitator function. J Bio Chem. 2011;286:19489–500.
Herz J. The LDL receptor gene family:(un) expected signal transducers in the brain. Neuron. 2001;29:571–81.
Kim N, Stiegler AL, Cameron TO, Hallock PT, Gomez AM, Huang JH, et al. Lrp4 is a receptor for Agrin and forms a complex with MuSK. Cell. 2008;135:334–42.
Zhang W, Coldefy AS, Hubbard SR, Burden SJ. Agrin binds to the N-terminal region of Lrp4 protein and stimulates association between Lrp4 and the first immunoglobulin-like domain in muscle-specific kinase (MuSK). J Bio Chem. 2011;286:40624–30.
Bao J, Zheng JJ, Wu D. The structural basis of DKK-mediated inhibition of Wnt/LRP signaling. Sci Signal. 2012;5:22.
Karner CM, Dietrich MF, Johnson EB, Kappesser N, Tennert C, Percin F, et al. Lrp4 regulates initiation of ureteric budding and is crucial for kidney formation–a mouse model for Cenani-Lenz syndrome. PloS One. 2010;5:e10418.
Glinka A, Wu W, Delius H, Monaghan AP, Blumenstock C, Niehrs C. Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature. 1998;391:357–62.
Semënov M, Tamai K, He X. SOST is a ligand for LRP5/LRP6 and a Wnt signaling inhibitor. J Bio Chem. 2005;280:26770–5.
Gong Y, Slee RB, Fukai N, Rawadi G, Roman-Roman S, Reginato AM, et al. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. 2001;107:513–23.
Clevers H. Wnt/β-catenin signaling in development and disease. Cell. 2006;127:469–80.
Acknowledgements
We highly appreciate participation of members of the families in the study presented here. Hammal Khan and Muhammad Bilal were supported by PhD fellowship awarded by Higher Education Commission (HEC), Islamabad, Pakistan. Angie Chong was supported by NUS Graduate Scholarship. Shifeng Xue was supported by NUS PYP.
Author information
Authors and Affiliations
Contributions
Hammal Khan and Muhammad Bilal performed the experimental work, analyzed the data, and prepared the manuscript. Shoaib Nawaz, Abdullah, Sanaullah Abbasi, Amir Hussain, and Shabir Hussain visited the families, drawn pedigrees, and collected blood samples. Angie Chong and Shifeng Xue performed the functional analysis. Imran Ullah and Wasim Ahmad designed the study, provided funds, and finalized the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Khan, H., Chong, A.E.Q., Bilal, M. et al. Novel variants in the LRP4 underlying Cenani-Lenz Syndactyly syndrome. J Hum Genet 67, 253–259 (2022). https://doi.org/10.1038/s10038-021-00995-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-021-00995-x
This article is cited by
-
New mutation in the β1 propeller domain of LRP4 responsible for congenital myasthenic syndrome associated with Cenani–Lenz syndrome
Scientific Reports (2023)
-
Intrafusal-fiber LRP4 for muscle spindle formation and maintenance in adult and aged animals
Nature Communications (2023)