Abstract
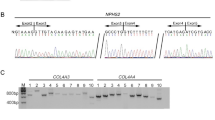
Cystic fibrosis is a hereditary disease that mostly affects the sweat glands, respiratory system, digestive system, and reproductive system. Many and various types of mutations have been reported in CFTR in different ethnicities and countries/regions. Analysis of CFTR gene rearrangements is recommended in patients with unidentified mutated alleles in CFTR sequencing analysis. We collected MLPA analyses of 527 patients from Turkey who had at least one unidentified mutation in CFTR sequence analysis. Heterozygous/homozygous deletions were detected in the CFTR gene in 49 individuals (9.2%) from 35 families. Twelve different single/multi exon deletions were demonstrated, two of which were not previously reported in the literature. Mutations have previously reported in patients from various regions including Asia, Europe, and Africa, and Turkey is located at a crossroads between them. The most frequent mutation was the exon 2 deletion, accounting for 60%. Moreover, patients with exon 2 deletions, were especially originated from northern Turkey. This finding is valuable in leading and shaping planned screening programs in Turkey. Our study, the most comprehensive study for rearrangement analysis in patients from Tukey, revealed a candidate hotspot region of patients suspected of having CFTR-related disorders from Turkey.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Bombieri C, Claustres M, De Boeck K, Derichs N, Dodge J, Girodon E, et al. Recommendations for the classification of diseases as CFTR-related disorders. J Cyst Fibros. 2011;10:S86–102.
Hamosh A, FitzSimmons SC, Macek M, Knowles MR, Rosenstein BJ, Cutting GR. Comparison of the clinical manifestations of cystic fibrosis in black and white patients. J Pediatr. 1998;132:255–9.
Mak V, Zielenski J, Tsui LC, Durie P, Zini A, Martin S, et al. Proportion of cystic fibrosis gene mutations not detected by routine testing in men with obstructive azoospermia. JAMA. 1999;281:2217–24.
Alper OM, Wong LJ, Young S, Pearl M, Graham S, Sherwin J, et al. Identification of novel and rare mutations in California Hispanic and African American cystic fibrosis patients. Hum Mutat. 2004;24:353.
Moskowitz SM, Chmiel JF, Sternen DL, Cheng E, Gibson RL, Marshall SG, et al. Clinical practice and genetic counseling for cystic fibrosis and CFTR-related disorders. Genet Med. 2008;10:851–68.
Derksen RH, Bouma BN, Kater L. The prevalence and clinical associations of the lupus anticoagulant in systemic lupus erythematosus. Scand J Rheumatol. 1987;16:185–92.
Ong T, Marshall SG, Karczeski BA, et al. Cystic Fibrosis and Congenital Absence of the Vas Deferens. 2001 Mar 26 [Updated 2017 Feb 2]. GeneReviews® [Internet].
Owczarzy R, Tataurov AV, Wu Y, Manthey JA, McQuisten KA, Almabrazi HG, et al. IDT SciTools: a suite for analysis and design of nucleic acid oligomers. Nucleic Acids Res. 2008;36:W163–9.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–8.
Dörk T, Macek M, Mekus F, Tümmler B, Tzountzouris J, Casals T, et al. Characterization of a novel 21-kb deletion, CFTRdele2,3(21 kb), in the CFTR gene: a cystic fibrosis mutation of Slavic origin common in Central and East Europe. Hum Genet. 2000;106:259–68.
Kilinç MO, Ninis VN, Dağli E, Demirkol M, Ozkinay F, Arikan Z, et al. Highest heterogeneity for cystic fibrosis: 36 mutations account for 75% of all CF chromosomes in Turkish patients. Am J Med Genet. 2002;113:250–7.
Lakeman P, Gille JJ, Dankert-Roelse JE, Heijerman HG, Munck A, Iron A, et al. CFTR mutations in Turkish and North African cystic fibrosis patients in Europe: implications for screening. Genet Test. 2008;12:25–35.
Onay T, Topaloglu O, Zielenski J, Gokgoz N, Kayserili H, Camcioglu Y, et al. Analysis of the CFTR gene in Turkish cystic fibrosis patients: identification of three novel mutations (3172delAC, P1013L and M1028I). Hum Genet. 1998;102:224–30.
Férec C, Casals T, Chuzhanova N, Macek M, Bienvenu T, Holubova A, et al. Gross genomic rearrangements involving deletions in the CFTR gene: characterization of six new events from a large cohort of hitherto unidentified cystic fibrosis chromosomes and meta-analysis of the underlying mechanisms. Eur J Hum Genet. 2006;14:567–76.
Niel F, Martin J, Dastot-Le Moal F, Costes B, Boissier B, Delattre V, et al. Rapid detection of CFTR gene rearrangements impacts on genetic counselling in cystic fibrosis. J Med Genet. 2004;41:e118.
Indika NLR, Vidanapathirana DM, Dilanthi HW, Kularatnam GAM, Chandrasiri NDPD, Jasinge E. Phenotypic spectrum and genetic heterogeneity of cystic fibrosis in Sri Lanka. BMC Med Genet. 2019;20:89.
Neocleous V, Yiallouros PK, Tanteles GA, Costi C, Moutafi M, Ioannou P, et al. Apparent homozygosity of p.Phe508del in CFTR due to a large gene deletion of exons 4-11. Case Rep Genet. 2014;2014:613863.
Chevalier-Porst F, Bonardot AM, Chazalette JP, Mathieu M, Bozon D. 40 kilobase deletion (CF 40 kb del 4-10) removes exons 4 to 10 of the cystic fibrosis transmembrane conductance regulator gene. Hum Mutat. 1998;S291-4.
Alibakhshi R, Kianishirazi R, Cassiman JJ, Zamani M, Cuppens H. Analysis of the CFTR gene in Iranian cystic fibrosis patients: identification of eight novel mutations. J Cyst Fibros. 2008;7:102–9.
Audrézet MP, Chen JM, Raguénès O, Chuzhanova N, Giteau K, Le Maréchal C, et al. Genomic rearrangements in the CFTR gene: extensive allelic heterogeneity and diverse mutational mechanisms. Hum Mutat. 2004;23:343–57.
Atag E, Bas Ikizoglu N, Ergenekon AP, Gokdemir Y, Eralp EE, Ata P, et al. Novel mutations and deletions in cystic fibrosis in a tertiary cystic fibrosis center in Istanbul. Pediatr Pulmonol. 2019;54:743–50.
Wakabayashi-Nakao K, Yu Y, Nakakuki M, Hwang TC, Ishiguro H, Sohma Y. Characterization of Δ(G970-T1122)-CFTR, the most frequent CFTR mutant identified in Japanese cystic fibrosis patients. J Physiol Sci. 2019;69:103–12.
Sohn YB, Ko JM, Jang JY, Seong MW, Park SS, Suh DI, et al. Deletion of exons 16-17b of CFTR is frequently identified in Korean patients with cystic fibrosis. Eur J Med Genet. 2019;62:103681.
Lerer I, Laufer-Cahana A, Rivlin JR, Augarten A, Abeliovich D. A large deletion mutation in the CFTR gene (3120+1Kbdel8.6Kb): a founder mutation in the Palestinian Arabs. Mutation in brief no. 231. Online. Hum Mutat. 1999;13:337.
Faà V, Bettoli PP, Demurtas M, Zanda M, Ferri V, Cao A, et al. A new insertion/deletion of the cystic fibrosis transmembrane conductance regulator gene accounts for 3.4% of cystic fibrosis mutations in Sardinia: implications for population screening. J Mol Diagn. 2006;8:499–503.
Kaplan S, Pinar G, Kaplan B, Aslantekin F, Karabulut E, Ayar B, et al. The prevalence of consanguineous marriages and affecting factors in Turkey: a national survey. J Biosoc Sci. 2016;48:616–30.
Claustres M, Thèze C. des Georges M, Baux D, Girodon E, Bienvenu T, et al. CFTR-France, a national relational patient database for sharing genetic and phenotypic data associated with rare CFTR variants. Hum Mutat. 2017;38:1297–315.
Chiang CWK, Marcus JH, Sidore C, Biddanda A, Al-Asadi H, Zoledziewska M, et al. Genomic history of the Sardinian population. Nat Genet. 2018;50:1426–34.
Elborn JS. Cystic fibrosis. Lancet. 2016;388:2519–31.
Acknowledgements
We thank the staff of the Genetic Diagnostic Center of Haseki Training and Research Hospital for their collaboration and Esranur Kucuktepe for her efforts in the validation of the experiments.
Funding
The authors received no financial support for the research, authorship, and/or publication of this article.
Author information
Authors and Affiliations
Contributions
MBD: Investigation, Formal analysis, Writing – original draft, Writing - review & editing, Project administration, Resources, and Supervision, POC: Investigation, Formal analysis, Methodology, and Writing.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The study protocol was conducted in accordance with the Declaration of Helsinki (1964) and was approved by ethics committee of Haseki Training and Research Hospital (ID:2020-76). Consent forms were obtained from all patients.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Duz, M.B., Ozyavuz Cubuk, P. Analysis of rearrangements of the CFTR gene in patients from Turkey with CFTR-related disorders: frequent exon 2 deletion. J Hum Genet 66, 315–320 (2021). https://doi.org/10.1038/s10038-020-00859-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-020-00859-w