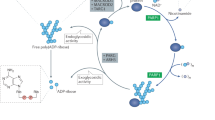
Abstract
Poly(ADP-ribose) polymerases (PARPs) modify target proteins post-translationally with poly(ADP-ribose) (PAR) or mono(ADP-ribose) (MAR) using NAD+ as substrate. The best-studied PARPs generate PAR modifications and include PARP1 and the tankyrase PARP5A, both of which are targets for cancer therapy with inhibitors in either clinical trials or preclinical development. There are 15 additional PARPs, most of which modify proteins with MAR, and their biology is less well understood. Recent data identify potentially cancer-relevant functions for these PARPs, which indicates that we need to understand more about these PARPs to effectively target them.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Rouleau, M., Patel, A., Hendzel, M., Kaufmann, S. & Poirier, G. PARP inhibition: PARP1 and beyond. Nature Rev. Cancer 10, 293–301 (2010).
Bürkle, A. Poly(ADP-ribose). The most elaborate metabolite of NAD+. FEBS J. 272, 4576–4589 (2005).
Kleine, H. et al. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol. Cell 32, 57–69 (2008).
Marsischky, G. T., Wilson, B. A. & Collier, R. J. Role of glutamic acid 988 of human poly-ADP-ribose polymerase in polymer formation. Evidence for active site similarities to the ADP-ribosylating toxins. J. Biol. Chem. 270, 3247–3254 (1995).
Otto, H. et al. In silico characterization of the family of PARP-like poly(ADP-ribosyl)transferases (pARTs). BMC Genom. 6, 139 (2005).
Wahlberg, E. et al. Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors. Nature Biotech. 30, 283–288 (2012).
Ruf, A., Rolli, V., de Murcia, G. & Schulz, G. The mechanism of the elongation and branching reaction of poly(ADP-ribose) polymerase as derived from crystal structures and mutagenesis. J. Mol. Biol. 278, 57–65 (1998).
Han, S. & Tainer, J. A. The ARTT motif and a unified structural understanding of substrate recognition in ADP-ribosylating bacterial toxins and eukaryotic ADP-ribosyltransferases. Int. J. Med. Microbiol. 291, 523–529 (2002).
D'Amours, D., Desnoyers, S., D'Silva, I. & Poirier, G. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 342, 249–268 (1999).
Alvarez-Gonzalez, R. & Jacobson, M. Characterization of polymers of adenosine diphosphate ribose generated in vitro and in vivo. Biochemistry 26, 3218–3224 (1987).
Sakura, H. et al. Natural occurence of a biopolymer, poly (adenosine diphosphate ribose). Nucleic Acids Res. 4, 2903–2915 (1977).
Affar, E. et al. Immunological determination and size characterization of poly(ADP-ribose) synthesized in vitro and in vivo. Biochim. Biophys. Acta 1428, 137–146 (1999).
Gibson, B. & Kraus, W. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nature Rev. Mol. Cell Biol. 13, 411–424 (2012).
Malanga, M. & Althaus, F. The role of poly(ADP-ribose) in the DNA damage signaling network. Biochem. Cell Biol. 83, 354–364 (2005).
Stilmann, M. et al. A nuclear poly(ADP-ribose)-dependent signalosome confers DNA damage-induced IκB kinase activation. Mol. Cell 36, 365–378 (2009).
Kotova, E., Jarnik, M. & Tulin, A. Poly (ADP-ribose) polymerase 1 is required for protein localization to Cajal body. PLoS genet. 5, e1000387 (2009).
Leung, A. et al. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol. Cell 42, 489–499 (2011).
Chang, P., Coughlin, M. & Mitchison, T. J. Interaction between Poly(ADP-ribose) and NuMA contributes to mitotic spindle pole assembly. Mol. Biol. Cell 20, 4575–4585 (2009).
Wang, Z. et al. Recognition of the iso-ADP-ribose moiety in poly(ADP-ribose) by WWE domains suggests a general mechanism for poly(ADP-ribosyl)ation-dependent ubiquitination. Genes Dev. 26, 235–240 (2012).
Ahel, I. et al. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature 451, 81–85 (2008).
Karras, G. et al. The macro domain is an ADP-ribose binding module. EMBO J. 24, 1911–1920 (2005).
Pleschke, J., Kleczkowska, H., Strohm, M. & Althaus, F. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem. 275, 40974–40980 (2000).
Barkauskaite, E., Jankevicius, G., Ladurner, A., Ahel, I. & Timinszky, G. The recognition and removal of cellular poly(ADP-ribose) signals. FEBS J. 280, 3491–3507 (2013).
Schultz, J., Milpetz, F., Bork, P. & Ponting, C. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl Acad. Sci. USA 95, 5857–5864 (1998).
Letunic, I., Doerks, T. & Bork, P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 40, 5 (2012).
Dani, N. et al. Combining affinity purification by ADP-ribose-binding macro domains with mass spectrometry to define the mammalian ADP-ribosyl proteome. Proc. Natl Acad. Sci. USA 106, 4243–4248 (2009).
Forst, A. et al. Recognition of mono-ADP-ribosylated ARTD10 substrates by ARTD8 macrodomains. Structure 21, 462–475 (2013).
Neuvonen, M. & Ahola, T. Differential activities of cellular and viral macro domain proteins in binding of ADP-ribose metabolites. J. Mol. Biol. 385, 212–225 (2009).
Feijs, K., Verheugd, P. & Lüscher, B. Expanding functions of intracellular resident mono-ADP-ribosylation in cell physiology. FEBS J. 280, 3519–3529 (2013).
Deng, Q. & Barbieri, J. Molecular mechanisms of the cytotoxicity of ADP-ribosylating toxins. Annu. Rev. Microbiol. 62, 271–288 (2008).
Tiemann, B. et al. ModA and ModB, two ADP-ribosyltransferases encoded by bacteriophage T4: catalytic properties and mutation analysis. J. Bacteriol. 186, 7262–7272 (2004).
Depping, R., Lohaus, C., Meyer, H. & Rüger, W. The mono-ADP-ribosyltransferases Alt and ModB of bacteriophage T4: target proteins identified. Biochem. Biophys. Res. Commun. 335, 1217–1223 (2005).
Haigis, M. et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic β cells. Cell 126, 941–954 (2006).
Liszt, G., Ford, E., Kurtev, M. & Guarente, L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J. Biol. Chem. 280, 21313–21320 (2005).
Glowacki, G. et al. The family of toxin-related ecto-ADP-ribosyltransferases in humans and the mouse. Protein Sci. 11, 1657–1670 (2002).
Terashima, M., Yamamori, C. & Shimoyama, M. ADP-ribosylation of Arg28 and Arg206 on the actin molecule by chicken arginine-specific ADP-ribosyltransferase. Eur. J. Biochem. 231, 242–249 (1995).
Terashima, M. et al. ADP-ribosylation of actins by arginine-specific ADP-ribosyltransferase purified from chicken heterophils. Eur. J. Biochem. 204, 305–311 (1992).
Huang, H., Graves, D., Robson, R. & Huiatt, T. ADP-ribosylation of the intermediate filament protein desmin and inhibition of desmin assembly in vitro by muscle ADP-ribosyltransferase. Biochem. Biophys. Res. Commun. 197, 570–577 (1993).
Gregory, H. L. & Barry, E. L. ADP-ribosylation of the 78-kDa glucose-regulated protein during nutritional stress. Eur. J. Biochem. 186, 205–211 (1989).
Laitusis, A., Brostrom, M. & Brostrom, C. The dynamic role of GRP78/BiP in the coordination of mRNA translation with protein processing. J. Biol. Chem. 274, 486–493 (1999).
Ledford, B. & Leno, G. ADP-ribosylation of the molecular chaperone GRP78/BiP. Mol. Cell. Biochem. 138, 141–148 (1994).
Chambers, J., Petrova, K., Tomba, G., Vendruscolo, M. & Ron, D. ADP ribosylation adapts an ER chaperone response to short-term fluctuations in unfolded protein load. J. Cell Biol. 198, 371–385 (2012).
Lupi, R., Corda, D. & Di Girolamo, M. Endogenous ADP-ribosylation of the G protein β subunit prevents the inhibition of type 1 adenylyl cyclase. J. Biol. Chem. 275, 9418–9424 (2000).
Lupi, R. et al. Endogenous mono-ADP-ribosylation of the free Gβγ prevents stimulation of phosphoinositide 3-kinase-gamma and phospholipase C-β2 and is activated by G-protein-coupled receptors. Biochem. J. 367, 825–832 (2002).
Bondarenko, V. A. et al. Residues within the polycationic region of cGMP phosphodiesterase γ subunit crucial for the interaction with transducin alpha subunit. Identification by endogenous ADP-rybosylation and site directed mutagenesis. J. Biol. Chem. 272, 15856–15864 (1997).
Bondarenko, V., Yamazaki, M., Hayashi, F. & Yamazaki, A. Suppression of GTP/T alpha-dependent activation of cGMP phosphodiesterase by ADP-ribosylation by its gamma subunit in amphibian rod photoreceptor membranes. Biochemistry 38, 7755–7763 (1999).
Dani, N. et al. Mono-ADP-ribosylation of the G protein βγ dimer is modulated by hormones and inhibited by Arf6. J. Biol. Chem. 286, 5995–6005 (2011).
Altmeyer, M., Messner, S., Hassa, P., Fey, M. & Hottiger, M. Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic Acids Res. 37, 3723–3738 (2009).
Messner, S. et al. PARP1 ADP-ribosylates lysine residues of the core histone tails. Nucleic Acids Res. 38, 6350–6362 (2010).
Tao, Z. Gao, P. & Liu, H.-w. Identification of the ADP-ribosylation sites in the PARP-1 automodification domain: analysis and implications. J. Am. Chem. Soc. 131, 14258–14260 (2009).
Jwa, M. & Chang, P. PARP16 is a tail-anchored endoplasmic reticulum protein required for the PERK- and IRE1α-mediated unfolded protein response. Nature Cell Biol. 14, 1223–1230 (2012).
Vyas, S., Chesarone-Cataldo, M., Todorova, T., Huang, Y. H. & Chang, P. A systematic analysis of the PARP protein family identifies new functions critical for cell physiology. Nature Commun. 4, 2240 (2013).
Luo, X. & Kraus, W. On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 26, 417–432 (2012).
Scarpa, E., Fabrizio, G. & Di Girolamo, M. A role of intracellular mono-ADP-ribosylation in cancer biology. FEBS J. 280, 3551–3562 (2013).
Dani, N., Barbosa, A. J., Del Rio, A. & Di Girolamo, M. ADP-ribosylated proteins as old and new drug targets for anticancer therapy: the example of ARF6. Curr. Pharm. Des. 19, 624–633 (2013).
Seo, G. et al. Reciprocal Inhibition between intracellular antiviral signaling and the RNAi machinery in mammalian cells. Cell Host Microbe 14, 435–445 (2013).
Verheugd, P. et al. Regulation of NF-κB signalling by the mono-ADP-ribosyltransferase ARTD10. Nature Commun. 4, 1683 (2013).
Barbarulo, A. et al. Poly(ADP-ribose) polymerase family member 14 (PARP14) is a novel effector of the JNK2-dependent pro-survival signal in multiple myeloma. Oncogene 32, 4231–4242 (2013).
Goenka, S. & Boothby, M. Selective potentiation of Stat-dependent gene expression by collaborator of Stat6 (CoaSt6), a transcriptional cofactor. Proc. Natl Acad. Sci. USA 103, 4210–4215 (2006).
Aguiar, R. et al. BAL is a novel risk-related gene in diffuse large B-cell lymphomas that enhances cellular migration. Blood 96, 4328–4334 (2000).
Kickhoefer, V. A. et al. The 193-kD vault protein, VPARP, is a novel poly(ADP-ribose) polymerase. J. Cell Biol. 146, 917–928 (1999).
Tuncel, H. et al. PARP6, a mono(ADP-ribosyl) transferase and a negative regulator of cell proliferation, is involved in colorectal cancer development. Int. J. Oncol. 41, 2079–2086 (2012).
Ma, Q., Baldwin, K., Renzelli, A., McDaniel, A. & Dong, L. TCDD-inducible poly(ADP-ribose) polymerase: a novel response to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Biochem. Biophys. Res. Commun. 289, 499–506 (2001).
MacPherson, L. et al. 2,3,7,8-Tetrachlorodibenzo-p-dioxin poly(ADP-ribose) polymerase (TiPARP, ARTD14) is a mono-ADP-ribosyltransferase and repressor of aryl hydrocarbon receptor transactivation. Nucleic Acids Res. 41, 1604–1621 (2013).
Walter, P. & Ron, D. The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086 (2011).
Tsai, Y. & Weissman, A. The unfolded protein response, degradation from endoplasmic reticulum and cancer. Genes Cancer 1, 764–778 (2010).
Atkins, C. et al. Characterization of a novel PERK kinase inhibitor with antitumor and antiangiogenic activity. Cancer Res. 73, 1993–2002 (2013).
Papandreou, I. et al. Identification of an Ire1α endonuclease specific inhibitor with cytotoxic activity against human multiple myeloma. Blood 117, 1311–1314 (2011).
Mimura, N. et al. Blockade of XBP1 splicing by inhibition of IRE1α is a promising therapeutic option in multiple myeloma. Blood 119, 5772–5781 (2012).
Di Paola, S., Micaroni, M., Di Tullio, G., Buccione, R. & Di Girolamo, M. PARP16/ARTD15 is a novel endoplasmic-reticulum-associated mono-ADP-ribosyltransferase that interacts with, and modifies karyopherin-ß1. PloS one 7, e37352 (2012).
Kauppinen, T. et al. Direct phosphorylation and regulation of poly(ADP-ribose) polymerase-1 by extracellular signal-regulated kinases 1/2. Proc. Natl Acad. Sci. USA 103, 7136–7141 (2006).
Cohen-Armon, M. et al. DNA-independent PARP-1 activation by phosphorylated ERK2 increases Elk1 activity: a link to histone acetylation. Mol. Cell 25, 297–308 (2007).
Yeh, T. Y., Sbodio, J. I. & Chi, N. W. Mitotic phosphorylation of tankyrase, a PARP that promotes spindle assembly, by GSK3. Biochem. Biophys. Res. Commun. 350, 574–579 (2006).
Patterson, J., Palombella, V., Fritz, C. & Normant, E. IPI-504, a novel and soluble HSP-90 inhibitor, blocks the unfolded protein response in multiple myeloma cells. Cancer Chemother. Pharmacol. 61, 923–932 (2008).
Obeng, E. et al. Proteasome inhibitors induce a terminal unfolded protein response in multiple myeloma cells. Blood 107, 4907–4916 (2006).
Anderson, P. & Kedersha, N. Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 33, 141–150 (2008).
Arimoto, K., Fukuda, H., Imajoh-Ohmi, S., Saito, H. & Takekawa, M. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nature Cell Biol. 10, 1324–1332 (2008).
Kathrin, T. et al. Inhibition of mTORC1 by astrin and stress granules prevents apoptosis in cancer cells. Cell 154, 859–874 (2013).
Wilson, W. & Hay, M. Targeting hypoxia in cancer therapy. Nature Rev. Cancer 11, 393–410 (2011).
Caravita, T., de Fabritiis, P., Palumbo, A., Amadori, S. & Boccadoro, M. Bortezomib: efficacy comparisons in solid tumors and hematologic malignancies. Nature Clin. Pract. Oncol. 3, 374–387 (2006).
Fournier, M.-J., Gareau, C. & Mazroui, R. The chemotherapeutic agent bortezomib induces the formation of stress granules. Cancer Cell. Int. 10, 12 (2010).
Lin, H. & Gregory, J. H. Correction: MicroRNAs: small RNAs with a big role in gene regulation. Nature Rev. Genet. 5, 522–531 (2004).
Li, L., Yu, C., Gao, H. & Li, Y. Argonaute proteins: potential biomarkers for human colon cancer. BMC Cancer 10, 38 (2010).
Cheng, N., Li, Y. & Han, Z.-G. Argonaute2 promotes tumor metastasis by way of up-regulating focal adhesion kinase expression in hepatocellular carcinoma. Hepatology 57, 1906–1918 (2013).
Benhamed, M., Herbig, U., Ye, T., Dejean, A. & Bischof, O. Senescence is an endogenous trigger for microRNA-directed transcriptional gene silencing in human cells. Nature Cell Biol. 14, 266–275 (2012).
Zhang, X., Graves, P. & Zeng, Y. Overexpression of human Argonaute2 inhibits cell and tumor growth. Biochim. Biophys. Acta 1830, 2553–2561 (2013).
Völler, D., Reinders, J., Meister, G. & Bosserhoff, A. K. Strong reduction of AGO2 expression in melanoma and cellular consequences. Br. J. Cancer 109, 3116–3124 (2013).
Yu, M. et al. PARP-10, a novel Myc-interacting protein with poly(ADP-ribose) polymerase activity, inhibits transformation. Oncogene 24, 1982–1993 (2005).
Vita, M. & Henriksson, M. The Myc oncoprotein as a therapeutic target for human cancer. Semin. Cancer Biol. 16, 318–330 (2006).
Chou, H.-Y. E., Chou, H. & Lee, S.-C. CDK-dependent activation of poly(ADP-ribose) polymerase member 10 (PARP10). J. Biol. Chem. 281, 15201–15207 (2006).
Herzog, N. et al. Caspase-dependent cleavage of the mono-ADP-ribosyltransferase ARTD10 interferes with its pro-apoptotic function. FEBS J. 280, 1330–1343 (2013).
Kleine, H. et al. Dynamic subcellular localization of the mono-ADP-ribosyltransferase ARTD10 and interaction with the ubiquitin receptor p62. Cell Commun. Signal. 10, 28 (2012).
Mathew, R., Karantza-Wadsworth, V. & White, E. Role of autophagy in cancer. Nature Rev. Cancer 7, 961–967 (2007).
Yang, Z. J., Chee, C. E., Huang, S. & Sinicrope, F. A. The role of autophagy in cancer: therapeutic implications. Mol. Cancer Ther. 10, 1533–1541 (2011).
Sui, X. et al. Autophagy and chemotherapy resistance: a promising therapeutic target for cancer treatment. Cell Death Dis. 4, e838 (2013).
Selvakumaran, M., Amaravadi, R., Vasilevskaya, I. & O'Dwyer, P. Autophagy inhibition sensitizes colon cancer cells to antiangiogenic and cytotoxic therapy. Clin. Cancer Res. 19, 2995–3007 (2013).
Yang, S. et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 25, 717–729 (2011).
Degenhardt, K. et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 10, 51–64 (2006).
Karantza-Wadsworth, V. et al. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 21, 1621–1635 (2007).
Trocoli, A. & Djavaheri-Mergny, M. The complex interplay between autophagy and NF-κB signaling pathways in cancer cells. Am. J. Cancer. Res. 1, 629–649 (2011).
Goenka, S., Cho, S. & Boothby, M. Collaborator of Stat6 (CoaSt6)-associated poly(ADP-ribose) polymerase activity modulates Stat6-dependent gene transcription. J. Biol. Chem. 282, 18732–18739 (2007).
Cho, S. et al. PARP-14, a member of the B aggressive lymphoma family, transduces survival signals in primary B cells. Blood 113, 2416–2425 (2009).
Cho, S. et al. Glycolytic rate and lymphomagenesis depend on PARP14, an ADP ribosyltransferase of the B aggressive lymphoma (BAL) family. Proc. Natl Acad. Sci. USA 108, 15972–15977 (2011).
Friedl, P. & Wolf, K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nature Rev. Cancer 3, 362–374 (2003).
Roussos, E., Condeelis, J. & Patsialou, A. Chemotaxis in cancer. Nature Rev. Cancer 11, 573–587 (2011).
Juszczynski, P. et al. BAL1 and BBAP are regulated by a gamma interferon-responsive bidirectional promoter and are overexpressed in diffuse large B-cell lymphomas with a prominent inflammatory infiltrate. Mol. Cell. Biol. 26, 5348–5359 (2006).
Wilkinson, P. & Islam, L. Recombinant IL-4 and IFN-γ activate locomotor capacity in human B lymphocytes. Immunology 67, 237–243 (1989).
Clinchy, B., Elenstrom, C. & Moller, G. The effect of T cell-derived cytokines on B cell motility in vitro. Cell. Immunol. 146, 62–70 (1993).
Zaidi, M. & Merlino, G. The two faces of interferon-γ in cancer. Clin. Cancer Res. 17, 6118–6124 (2011).
Champelovier, P. et al. Is interferon γ one key of metastatic potential increase in human bladder carcinoma? Clin. Cancer Res. 9, 4562–4569 (2003).
Lollini, P. et al. Inhibition of tumor growth and enhancement of metastasis after transfection of the γ-interferon gene. Int. J. Cancer 55, 320–329 (1993).
Bernabei, P. et al. Interferon-gamma receptor 2 expression as the deciding factor in human T, B, and myeloid cell proliferation or death. J. Leukocyte Biol. 70, 950–960 (2001).
Aguiar, R., Takeyama, K., He, C., Kreinbrink, K. & Shipp, M. B-aggressive lymphoma family proteins have unique domains that modulate transcription and exhibit poly(ADP-ribose) polymerase activity. J. Biol. Chem. 280, 33756–33765 (2005).
Kuo, J.-C., Han, X., Hsiao, C.-T., Yates, J. & Waterman, C. Analysis of the myosin-II-responsive focal adhesion proteome reveals a role for β-Pix in negative regulation of focal adhesion maturation. Nature Cell Biol. 13, 383–393 (2011).
Reticker-Flynn, N. et al. A combinatorial extracellular matrix platform identifies cell-extracellular matrix interactions that correlate with metastasis. Nature Commun. 3, 1122 (2012).
Akiyama, S., Olden, K. & Yamada, K. Fibronectin and integrins in invasion and metastasis. Cancer Metastasis Rev. 14, 173–189 (1995).
Penning, T. et al. Optimization of phenyl-substituted benzimidazole carboxamide poly(ADP-ribose) polymerase inhibitors: identification of (S)-2-(2-fluoro-4-(pyrrolidin-2-yl)phenyl)-1H-benzimidazole-4-carboxamide (A-966492), a highly potent and efficacious inhibitor. J. Med. Chem. 53, 3142–3153 (2010).
Alice, N. W. & Eddy, S. Y. Beyond, DNA repair: additional functions of PARP-1 in cancer. Front. Oncol. 3, 290 (2013).
Petesch, S. & Lis, J. Activator-induced spread of poly(ADP-ribose) polymerase promotes nucleosome loss at Hsp70. Mol. Cell 45, 64–74 (2012).
Amé, J. et al. PARP-2, A novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J. Biol. Chem. 274, 17860–17868 (1999).
Loseva, O. et al. PARP-3 is a mono-ADP-ribosylase that activates PARP-1 in the absence of DNA. J. Biol. Chem. 285, 8054–8060 (2010).
Boehler, C. et al. Poly(ADP-ribose) polymerase 3 (PARP3), a newcomer in cellular response to DNA damage and mitotic progression. Proc. Natl Acad. Sci. USA 108, 2783–2788 (2011).
Smith, S., Giriat, I., Schmitt, A. & de Lange, T. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science 282, 1484–1487 (1998).
Smith, S. & de Lange, T. Tankyrase promotes telomere elongation in human cells. Curr. Biol. 10, 1299–1302 (2000).
Huang, S.-M. A. et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 461, 614–620 (2009).
Cho-Park, P. & Steller, H. Proteasome regulation by ADP-ribosylation. Cell 153, 614–627 (2013).
Chang, P., Coughlin, M. & Mitchison, T. Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nature Cell Biol. 7, 1133–1139 (2005).
Cook, B., Dynek, J., Chang, W., Shostak, G. & Smith, S. Role for the related poly(ADP-Ribose) polymerases tankyrase 1 and 2 at human telomeres. Mol. Cell. Biol. 22, 332–342 (2002).
Acknowledgements
The authors thank F. Solomon, M. Vander Heiden, J. Pascal and F. Bock for their helpful comments and discussion about the manuscript. This work is supported by a grant to P.C. from the US National Institutes of Health (ROI GM087465) and was partially supported by Cancer Center Support (core; grant P30-CA14051).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
DATABASES
Rights and permissions
About this article
Cite this article
Vyas, S., Chang, P. New PARP targets for cancer therapy. Nat Rev Cancer 14, 502–509 (2014). https://doi.org/10.1038/nrc3748
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrc3748
This article is cited by
-
Crosstalk between protein post-translational modifications and phase separation
Cell Communication and Signaling (2024)
-
Role of poly(ADP-ribose) polymerase-1 in regulating human islet cell differentiation
Scientific Reports (2022)
-
Recent advances in structural types and medicinal chemistry of PARP-1 inhibitors
Medicinal Chemistry Research (2022)
-
Small molecules in targeted cancer therapy: advances, challenges, and future perspectives
Signal Transduction and Targeted Therapy (2021)
-
HPF1 remodels the active site of PARP1 to enable the serine ADP-ribosylation of histones
Nature Communications (2021)