Abstract
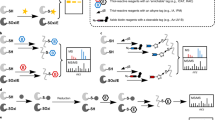
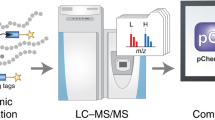
Chemical probes that target classes of proteins based on shared functional properties have emerged as powerful tools for proteomics. The metabolome rivals, if not surpasses, the proteome in terms of size and complexity, suggesting that efforts to profile metabolites would also benefit from targeted technologies. Here we apply the principle of chemoselective probes to the metabolome, creating a general strategy to tag, enrich and profile large classes of small molecules from biological systems. Key to success was incorporation of a protease-cleavage step to release captured metabolites in a format compatible with liquid chromatography–mass spectrometry (LC-MS) analysis. This technology, termed metabolite enrichment by tagging and proteolytic release (METPR), is applicable to small molecules of any physicochemical class, including polar, labile and low-mass (<100 Da) compounds. We applied METPR to profile changes in the thiol metabolome of human cancer cells treated with the antioxidant N-acetyl-L-cysteine.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Brown, P.O. & Botstein, D. Exploring the new world of the genome with DNA microarrays. Nat. Genet. 21, 33–37 (1999).
Patterson, S.D. & Aebersold, R. Proteomics: the first decade and beyond. Nat. Genet. 33, 311–323 (2003).
Fiehn, O. Metabolomics—the link between genotypes and phenotypes. Plant Mol. Biol. 48, 155–171 (2002).
Saghatelian, A. & Cravatt, B.F. Global strategies to integrate the proteome and metabolome. Curr. Opin. Chem. Biol. 9, 62–68 (2005).
Kell, D.B. & Westerhoff, H.V. Towards a rationale approach to the optimization of flux in microbial biotransformations. Trends Biotechnol. 4, 137–142 (1986).
Fell, D.A. Enzymes, metabolites and fluxes. J. Exp. Bot. 56, 267–272 (2005).
Fernie, A.R., Trethewey, R.N., Krotzky, A.J. & Willmitzer, L. Metabolite profiling: from diagnostics to systems biology. Nat. Rev. Mol. Cell Biol. 5, 763–769 (2004).
Wenk, M.R. The emerging field of lipidomics. Nat. Rev. Drug Discov. 4, 594–610 (2005).
Saghatelian, A. et al. Assignment of endogenous substrates to enzymes by global metabolite profiling. Biochemistry 43, 14332–14339 (2004).
Chiang, K.P., Niessen, S., Saghatelian, A. & Cravatt, B.F. An enzyme that regulates ether lipid signaling pathways in cancer annotated by multidimensional profiling. Chem. Biol. 13, 1041–1050 (2006).
Speers, A.E. & Cravatt, B.F. Chemical strategies for activity-based proteomics. ChemBioChem 5, 41–47 (2004).
Evans, M.J. & Cravatt, B.F. Mechanism-based profiling of enzyme families. Chem. Rev. 106, 3279–3301 (2006).
Adam, G.C., Sorensen, E.J. & Cravatt, B.F. Chemical strategies for functional proteomics. Mol. Cell. Proteomics 1, 781–790 (2002).
Zhang, H., Yan, W. & Aebersold, R. Chemical probes and tandem mass spectrometry: a strategy for the quantitative analysis of proteomes and subproteomes. Curr. Opin. Chem. Biol. 8, 66–75 (2004).
Daykin, C.A., Foxall, P.J.D., Connor, S.C., Lindon, J.C. & Nicholson, J.K. The comparison of plasma deproteinization methods for the detection of low-molecular-weight metabolites by (1)H nuclear magnetic resonance spectroscopy. Anal. Biochem. 304, 220–230 (2002).
Want, E.J. et al. Solvent-dependent metabolite distribution, clustering, and protein extraction for serum profiling with mass spectrometry. Anal. Chem. 78, 743–752 (2006).
Chowdhury, S.M., Munske, G.R., Siems, W.F. & Bruce, J.E. A new maleimide-bound acid-cleavable solid-support reagent for profiling phosphorylation. Rapid Commun. Mass Spectrom. 19, 899–909 (2005).
Tumelty, D., Cao, K. & Holmes, C.P. Traceless solid-phase synthesis of substituted benzimidazoles via a base-cleavable linker. Org. Lett. 3, 83–86 (2001).
Williams, S.J., Hekmat, O. & Withers, S.G. Synthesis and testing of mechanism-based protein-profiling probes for retaining endo-glycosidases. ChemBioChem 7, 116–124 (2006).
Smith, C.A., Want, E.J., O'Maille, G., Abagyan, R. & Siuzdak, G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 78, 779–787 (2006).
Roessner-Tunali, U. et al. Metabolic profiling of transgenic tomato plants overexpressing hexokinase reveals that the influence of hexose phosphorylation diminishes during fruit development. Plant Physiol. 133, 84–99 (2003).
Kell, D.B. Metabolomics and systems biology: making sense of the soup. Curr. Opin. Microbiol. 7, 296–307 (2004).
Yalcin, T. & Harrison, A.G. Ion chemistry of protonated lysine derivatives. J. Mass Spectrom. 31, 1237–1243 (1996).
Menon, S.G., Coleman, M.C., Walsh, S.A., Spitz, D.R. & Goswami, P.C. Differential susceptibility of nonmalignant human breast epithelial cells and breast cancer cells to thiol antioxidant-induced G(1)-delay. Antioxid. Redox Signal. 7, 711–718 (2005).
Shibanuma, M., Kuroki, T. & Nose, K. Induction of DNA-replication and expression of proto-oncogene c-myc and c-fos in quiescent Balb/3T3 cells by xanthine-xanthine oxidase. Oncogene 3, 17–21 (1988).
Kim, K.Y., Rhim, T., Choi, I. & Kim, S.S. N-acetylcysteine induces cell cycle arrest in hepatic stellate cells through its reducing activity. J. Biol. Chem. 276, 40591–40598 (2001).
Menon, S.G. et al. Redox regulation of the G1 to S phase transition in the mouse embryo fibroblast cell cycle. Cancer Res. 63, 2109–2117 (2003).
Bajad, S.U. et al. Separation and quantitation of water soluble cellular metabolites by hydrophilic interaction chromatography-tandem mass spectrometry. J. Chromatogr. A. 1125, 76–88 (2006).
Breitling, R., Pitt, A.R. & Barrett, M.P. Precision mapping of the metabolome. Trends Biotechnol. 24, 543–548 (2006).
Hemstrom, P. & Irgum, K. Hydrophilic interaction chromatography. J. Sep. Sci. 29, 1784–1821 (2006).
Acknowledgements
We thank H.P. Benton and A. Nordström for assistance with Q-TOF experiments. We also thank G. Simon for help in generating figures, and members of the Cravatt laboratory for helpful discussion and critical review of the manuscript. This work was supported by the American Cancer Society (PF-06-009-01-CDD, to E.E.C.), the National Institutes of Health (CA087660) and the Skaggs Institute for Chemical Biology.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Synthesis of reactive group-functionalized resins. (PDF 124 kb)
Supplementary Fig. 2
Protein precipitation as assessed by SDS-PAGE (10% SDS) visualized with silver stain. (PDF 129 kb)
Supplementary Fig. 3
Profiling metabolites with varied physicochemical properties by METPR. (PDF 154 kb)
Supplementary Fig. 4
METPR analysis of small molecule standards in human urine. (PDF 103 kb)
Supplementary Fig. 5
MS2 analysis of tagged small-molecules generated in METPR experiments. (PDF 191 kb)
Supplementary Table 1
Table of detected masses from NAC treatment. (PDF 43 kb)
Rights and permissions
About this article
Cite this article
Carlson, E., Cravatt, B. Chemoselective probes for metabolite enrichment and profiling. Nat Methods 4, 429–435 (2007). https://doi.org/10.1038/nmeth1038
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth1038
This article is cited by
-
Chemoselective Probes Serving as Promising Derivatization Tools in Targeted Metabolomics Research
Journal of Analysis and Testing (2020)
-
Isotopic Exchange HPLC-HRMS/MS Applied to Cyclic Proanthocyanidins in Wine and Cranberries
Journal of the American Society for Mass Spectrometry (2018)
-
Plasmonic silver nanoshells for drug and metabolite detection
Nature Communications (2017)
-
Chemistry Methods
Nature Methods (2011)
-
Database Resources in Metabolomics: An Overview
Journal of Neuroimmune Pharmacology (2010)